
Contributions
Abstract: EP960
Type: E-Poster Presentation
Session title: Myeloma and other monoclonal gammopathies - Biology & Translational Research
Background
Multiple myeloma remains incurable mostly due to acquired resistance. Understanding the mechanisms of resistance would allow to create an algorithm for selecting optimal therapies and pave the road to new treatments.
Aims
Since proteomic and genomic analysis of myeloma cells did not reveal therapeutically relevant findings, we applied metabolomics to analyze biological alterations that allow for acquisition of resistance to carfilzomib – widely used proteasome inhibitor.
Methods
GC-MS-based comparative metabolomics was applied to compare the primary carfilzomib sensitive myeloma cell lines AMO1 and RPMI (RPMI 8226) with their carfilzomib resistant progenies generated by prolonged carfilzomib exposure. Cells were lysed, fractionated metabolite extracts were derivatized and submitted to GC/MS analysis performed with Agilent 7890A GCxGC (Agilent Technologies) combined with Pegasus 4D TOF-MS (Leco). MS data were normalized to total ion current (TIC) and analyzed using Perseus and MetaboAnalyst. The significance was estimated by the two-sample T-test and one-way ANOVA with FDR correction. Multivariate analyses were carried out by untargeted principal component analysis (PCA) and hierarchical clustering. Differentially accumulated metabolites (DAMs) were subjected to the MetaboAnalyst and Ingenuity Pathway Analysis software to determine canonical pathways, biological functions and upstream regulators.
Results
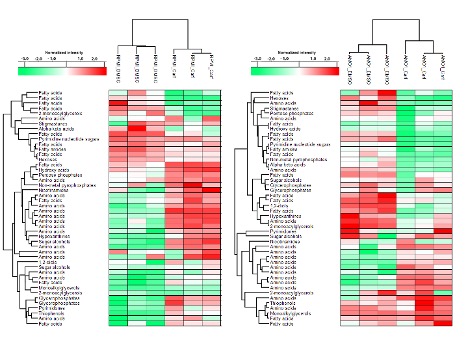
PCA revealed significant differences between sensitive and resistant progenies. 24 and 34 DAMs were identified in AMO1- and RPMI-resistant (Fig. 1) cells respectively, as compared to primary cells, 16 DAMs were common for both comparisons. The top-ranked canonical pathways included: aminoacyl-tRNA biosynthesis, amino acid’s metabolism and biosynthesis of unsaturated fatty acids.
The analysis of functions of DAMs revealed overrepresented categories associated with oxidation of glucose-6-phosphate (p 1.6E-08 for AMO1, 1.84E-08 for RPMI) and accumulation of lipids (p 1.39E-05 for AMO1, 5.97E-08 for RPMI) - the oxidation of glucose-6-phosphate was activated (z-score 2.86 for RPMI and 2.00 for AMO1), while accumulation of lipids was inhibited in resistant cells (z-scores -2.37 for AMOA1, -1.91 for RPMI).
Network analysis of DAMs in AMO1 revealed that PML transcription regulator was predicted to be the top positive regulator, while ARNT (Aryl hydrocarbon receptor nuclear translocator) and D-glucose were predicted to be the negative regulators in resistant cells (p 4.49E-05 and 9.46E-06, z-scores: 2 and 2.23). The DAMs negatively regulated by ARNT and D-glucose and positively regulated by PML belonged to processes related to lipids accumulation, acylglycerol and pyruvic acid, molecules down-regulated in AMO1 resistant cells. Downregulation of these compounds induces an oxidation of glucose-6-phosphate in resistant cells. In RPMI, UCP1 (Uncoupling protein 1) transporter was identified as the positive upstream regulator of observed differences in resistant cells and functionally connected to lipid accumulation and D-glucose level. It might suggest that increased level of this regulator can lead to observed inhibition of lipids accumulation.
Conclusion
We successfully applied metabolomics to assess biological alterations in myeloma resistant cells. Our results are in line with previous studies underlining the role of glucose and fatty acids metabolism in acquired resistance in myeloma. Confirmation of results in clinical setting is needed for potential development of new drugs targeting disrupted metabolic pathways to overcome resistance.
Keyword(s): Myeloma, Proteasome inhibitor, Resistance
Abstract: EP960
Type: E-Poster Presentation
Session title: Myeloma and other monoclonal gammopathies - Biology & Translational Research
Background
Multiple myeloma remains incurable mostly due to acquired resistance. Understanding the mechanisms of resistance would allow to create an algorithm for selecting optimal therapies and pave the road to new treatments.
Aims
Since proteomic and genomic analysis of myeloma cells did not reveal therapeutically relevant findings, we applied metabolomics to analyze biological alterations that allow for acquisition of resistance to carfilzomib – widely used proteasome inhibitor.
Methods
GC-MS-based comparative metabolomics was applied to compare the primary carfilzomib sensitive myeloma cell lines AMO1 and RPMI (RPMI 8226) with their carfilzomib resistant progenies generated by prolonged carfilzomib exposure. Cells were lysed, fractionated metabolite extracts were derivatized and submitted to GC/MS analysis performed with Agilent 7890A GCxGC (Agilent Technologies) combined with Pegasus 4D TOF-MS (Leco). MS data were normalized to total ion current (TIC) and analyzed using Perseus and MetaboAnalyst. The significance was estimated by the two-sample T-test and one-way ANOVA with FDR correction. Multivariate analyses were carried out by untargeted principal component analysis (PCA) and hierarchical clustering. Differentially accumulated metabolites (DAMs) were subjected to the MetaboAnalyst and Ingenuity Pathway Analysis software to determine canonical pathways, biological functions and upstream regulators.
Results
PCA revealed significant differences between sensitive and resistant progenies. 24 and 34 DAMs were identified in AMO1- and RPMI-resistant (Fig. 1) cells respectively, as compared to primary cells, 16 DAMs were common for both comparisons. The top-ranked canonical pathways included: aminoacyl-tRNA biosynthesis, amino acid’s metabolism and biosynthesis of unsaturated fatty acids.
The analysis of functions of DAMs revealed overrepresented categories associated with oxidation of glucose-6-phosphate (p 1.6E-08 for AMO1, 1.84E-08 for RPMI) and accumulation of lipids (p 1.39E-05 for AMO1, 5.97E-08 for RPMI) - the oxidation of glucose-6-phosphate was activated (z-score 2.86 for RPMI and 2.00 for AMO1), while accumulation of lipids was inhibited in resistant cells (z-scores -2.37 for AMOA1, -1.91 for RPMI).
Network analysis of DAMs in AMO1 revealed that PML transcription regulator was predicted to be the top positive regulator, while ARNT (Aryl hydrocarbon receptor nuclear translocator) and D-glucose were predicted to be the negative regulators in resistant cells (p 4.49E-05 and 9.46E-06, z-scores: 2 and 2.23). The DAMs negatively regulated by ARNT and D-glucose and positively regulated by PML belonged to processes related to lipids accumulation, acylglycerol and pyruvic acid, molecules down-regulated in AMO1 resistant cells. Downregulation of these compounds induces an oxidation of glucose-6-phosphate in resistant cells. In RPMI, UCP1 (Uncoupling protein 1) transporter was identified as the positive upstream regulator of observed differences in resistant cells and functionally connected to lipid accumulation and D-glucose level. It might suggest that increased level of this regulator can lead to observed inhibition of lipids accumulation.

Conclusion
We successfully applied metabolomics to assess biological alterations in myeloma resistant cells. Our results are in line with previous studies underlining the role of glucose and fatty acids metabolism in acquired resistance in myeloma. Confirmation of results in clinical setting is needed for potential development of new drugs targeting disrupted metabolic pathways to overcome resistance.
Keyword(s): Myeloma, Proteasome inhibitor, Resistance


