
Contributions
Abstract: EP936
Type: E-Poster Presentation
Session title: Myeloma and other monoclonal gammopathies - Biology & Translational Research
Background
The BOSTON study, a phase 3, randomized, open label clinical trial, has shown a clinical benefit of selinexor combined with bortezomib and dexamethasone (XVd) in patients with multiple myeloma. Selinexor acts by inhibiting the nuclear export protein XPO1, however, XPO1 expression does not correlate with response and the biological mechanisms underlying treatment response remain unclear. Additionally, while the overall response rate for XVd in the BOSTON study was 76.4%, there are currently no known genomic biomarkers to help guide treatment recommendations.
Aims
In this study, we characterized genomic and transcriptomic correlates of response to selinexor and identified potential biomarkers for clinical use.
Methods
We performed RNA sequencing on CD138+ cells from 100 patients who participated in the BOSTON study. We performed differential expression, followed by pathway analysis, to compare patients with long and short progression-free survival (PFS) in the XVd arm of the BOSTON dataset. Using the differentially expressed genes, we calculated GSVA (Gene Set Variation Analysis) scores and performed time-to-event univariate Cox proportional hazard models and log-rank testing to identify a novel gene expression signature that predicts PFS in the BOSTON dataset. We validated our signature using transcriptomic data from 64 patients from STORM, a clinical trial of selinexor combined with dexamethasone in patients with penta-refractory multiple myeloma.
Results
Through gene expression analysis, we identified a cluster of patients who are more likely to experience longer PFS with XVd as opposed to Vd (log rank p = 0.012, Fig 1A-B). We found that patients within the XVd arm in this cluster tend to be younger (wilcoxon p = 0.0029). Within this cluster of responders, we identified a total of 198 unique genes associated with longer PFS in the XVd arm (FDR < 0.05) that were not significantly associated with PFS in the Vd group. Furthermore, pathway analysis revealed increased interferon signaling, Myc deactivation, and proteasome downregulation in patients with longer PFS in this cluster (FDR < 0.05, Fig 1B). We identified a GSVA-based expression signature consisting of five genes, ASTN1, REC114, CIART, RPL23AP65, and NPHS1, that were downregulated in XVd patients with PFS > 300 days (Fig 1C). Using this signature, we accurately distinguished patients with long term PFS in the XVd arm of the BOSTON study (Cox proportional hazard model, FDR=0.003, hazard ratio=6.7, Fig 1D). Three of these five genes, ASTN1, NPHS1, and RPL23AP65, were also downregulated in patients that had better depth of response, defined as very good partial response (VGPR) or better. This signature validated successfully in the STORM study (Cox proportional hazard model, p=0.0095, hazard ratio=2, Fig 1D).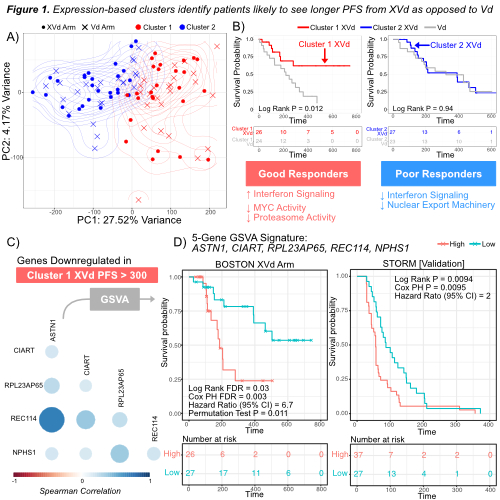
Conclusion
We report a novel, prognostic biomarker signature for selinexor, bortezomib and dexamethasone response in patients with multiple myeloma. We have validated our findings in the STORM cohort and are currently validating our findings in additional, independent cohorts of multiple myeloma patients treated with selinexor. This signature has important clinical relevance as it could identify patients most likely to benefit from treatment with selinexor-based therapy.
Keyword(s): Drug sensitivity, Gene expression profile, Multiple myeloma, Survival prediction
Abstract: EP936
Type: E-Poster Presentation
Session title: Myeloma and other monoclonal gammopathies - Biology & Translational Research
Background
The BOSTON study, a phase 3, randomized, open label clinical trial, has shown a clinical benefit of selinexor combined with bortezomib and dexamethasone (XVd) in patients with multiple myeloma. Selinexor acts by inhibiting the nuclear export protein XPO1, however, XPO1 expression does not correlate with response and the biological mechanisms underlying treatment response remain unclear. Additionally, while the overall response rate for XVd in the BOSTON study was 76.4%, there are currently no known genomic biomarkers to help guide treatment recommendations.
Aims
In this study, we characterized genomic and transcriptomic correlates of response to selinexor and identified potential biomarkers for clinical use.
Methods
We performed RNA sequencing on CD138+ cells from 100 patients who participated in the BOSTON study. We performed differential expression, followed by pathway analysis, to compare patients with long and short progression-free survival (PFS) in the XVd arm of the BOSTON dataset. Using the differentially expressed genes, we calculated GSVA (Gene Set Variation Analysis) scores and performed time-to-event univariate Cox proportional hazard models and log-rank testing to identify a novel gene expression signature that predicts PFS in the BOSTON dataset. We validated our signature using transcriptomic data from 64 patients from STORM, a clinical trial of selinexor combined with dexamethasone in patients with penta-refractory multiple myeloma.
Results
Through gene expression analysis, we identified a cluster of patients who are more likely to experience longer PFS with XVd as opposed to Vd (log rank p = 0.012, Fig 1A-B). We found that patients within the XVd arm in this cluster tend to be younger (wilcoxon p = 0.0029). Within this cluster of responders, we identified a total of 198 unique genes associated with longer PFS in the XVd arm (FDR < 0.05) that were not significantly associated with PFS in the Vd group. Furthermore, pathway analysis revealed increased interferon signaling, Myc deactivation, and proteasome downregulation in patients with longer PFS in this cluster (FDR < 0.05, Fig 1B). We identified a GSVA-based expression signature consisting of five genes, ASTN1, REC114, CIART, RPL23AP65, and NPHS1, that were downregulated in XVd patients with PFS > 300 days (Fig 1C). Using this signature, we accurately distinguished patients with long term PFS in the XVd arm of the BOSTON study (Cox proportional hazard model, FDR=0.003, hazard ratio=6.7, Fig 1D). Three of these five genes, ASTN1, NPHS1, and RPL23AP65, were also downregulated in patients that had better depth of response, defined as very good partial response (VGPR) or better. This signature validated successfully in the STORM study (Cox proportional hazard model, p=0.0095, hazard ratio=2, Fig 1D).
Conclusion
We report a novel, prognostic biomarker signature for selinexor, bortezomib and dexamethasone response in patients with multiple myeloma. We have validated our findings in the STORM cohort and are currently validating our findings in additional, independent cohorts of multiple myeloma patients treated with selinexor. This signature has important clinical relevance as it could identify patients most likely to benefit from treatment with selinexor-based therapy.
Keyword(s): Drug sensitivity, Gene expression profile, Multiple myeloma, Survival prediction


