
Contributions
Abstract: EP906
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Systemic inflammatory and auto-immune diseases (SIAD), often including relapsing polychondritis (RP), are observed in 25% of patients with MDS or CMML, although the causal relationship is unclear. We previously reported in 85 MDS/CMML patients with SIAD (Zhao et al, Leukemia 2021) that TET2, IDH1/2 and SRSF2 mutations were more frequent in those patients compared to a control cohort of MDS/CMML patients without SIAD, and correlated with naïve/memory T cell imbalance and defects in immune checkpoint receptors expression.
Recently, somatic mutations in the UBA1 gene were described, defining a novel disease called VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic) syndrome (Beck et al, NEJM 2020). 24% of the patients with VEXAS syndrome were reported to have MDS, and 60% had a previous diagnosis of RP.
Aims
To explore the prevalence of UBA1 mutations in patients diagnosed with MDS/CMML and SIAD
Methods
From our cohort of 85 patients with MDS/CMML and SIAD, all male patients (n=45) for whom material was available (n=33) were analyzed for the presence of UBA1 mutations. DNA was obtained from peripheral blood or bone marrow mononuclear cells and Sanger sequencing of exons 2 and 3 as well as exon-intron junctions was performed.
Results
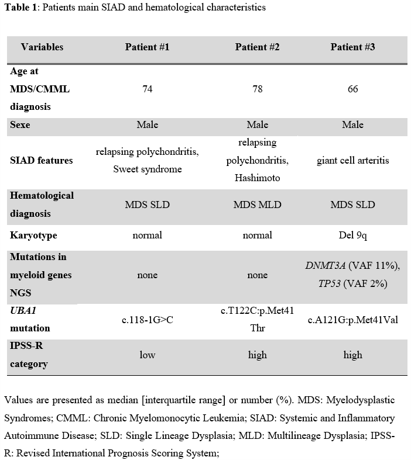
The 33 patients with SIAD included 6 MDS with excess blasts, 21 MDS without excess blasts and 6 CMML. 3 of them had UBA1 mutations. All 3 patients had MDS without excess of blasts, and no CMML patient had UBA1 mutation. Review of their bone marrow smears showed numerous vacuoles in myeloid and erythroid precursors, which were however not specific of UBA1 mutated cases of MDS. Patients #1 and #2 had normal karyotype while patient #3 had 9q deletion (Table 1). Mutations in UBA1 affected methionine-41 in patient #2 and patient #3 as reported by Beck et al, while patient #1 had a newly described mutation of a splice site predicted to have similar consequence on UBA1 protein (Bourbon et al. Blood 2021). Patient #3 had also somatic mutations in DNMT3A and TP53 (VAF 11% and 2%, respectively), while no other mutation was detected in patients #1 and #2 in our next-generation sequencing panel including 80 genes involved in myeloid malignancies.
3 patients in our MDS/CMML SIAD cohort had relapsing polychondritis, and 2 of them (patients #1 and #2) had UBA1 mutations. The last patient with UBA1 mutation (#3) had giant cell arteritis (GCA) (out of 7 GCA diagnosis in our SIAD cohort). All 3 patients with UBA1 mutation had steroid refractory SIAD and were heavily treated with other immunosuppressive therapies including methotrexate, azathioprine or cytokine targeting agents. Patients #1 and #3 received azacytidine (AZA) for their SIAD, based on our previous experience (Fraison et al. Leukemia Research 2016), with a partial response allowing dose reduction of steroids. At last follow up, none of the 3 patients with UBA1 mutation had MDS progression, and patient #2 died from a stroke 2.4 years after MDS diagnosis.
Conclusion
Our retrospective study represents the first assessment of UBA1 mutation among patients with myeloid malignancies associated with SIAD. The low prevalence of UBA1 mutations in our cohort suggests that other pathophysiological mechanisms may drive inflammation in a majority of MDS/CMML patients with associated SIAD, including mutations in epigenetic regulators TET2/IDH and SRSF2 as we previously proposed.
Keyword(s): Chronic myelomonocytic leukemia, Genetic, Immunology, Myelodysplasia
Abstract: EP906
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Systemic inflammatory and auto-immune diseases (SIAD), often including relapsing polychondritis (RP), are observed in 25% of patients with MDS or CMML, although the causal relationship is unclear. We previously reported in 85 MDS/CMML patients with SIAD (Zhao et al, Leukemia 2021) that TET2, IDH1/2 and SRSF2 mutations were more frequent in those patients compared to a control cohort of MDS/CMML patients without SIAD, and correlated with naïve/memory T cell imbalance and defects in immune checkpoint receptors expression.
Recently, somatic mutations in the UBA1 gene were described, defining a novel disease called VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic) syndrome (Beck et al, NEJM 2020). 24% of the patients with VEXAS syndrome were reported to have MDS, and 60% had a previous diagnosis of RP.
Aims
To explore the prevalence of UBA1 mutations in patients diagnosed with MDS/CMML and SIAD
Methods
From our cohort of 85 patients with MDS/CMML and SIAD, all male patients (n=45) for whom material was available (n=33) were analyzed for the presence of UBA1 mutations. DNA was obtained from peripheral blood or bone marrow mononuclear cells and Sanger sequencing of exons 2 and 3 as well as exon-intron junctions was performed.
Results
The 33 patients with SIAD included 6 MDS with excess blasts, 21 MDS without excess blasts and 6 CMML. 3 of them had UBA1 mutations. All 3 patients had MDS without excess of blasts, and no CMML patient had UBA1 mutation. Review of their bone marrow smears showed numerous vacuoles in myeloid and erythroid precursors, which were however not specific of UBA1 mutated cases of MDS. Patients #1 and #2 had normal karyotype while patient #3 had 9q deletion (Table 1). Mutations in UBA1 affected methionine-41 in patient #2 and patient #3 as reported by Beck et al, while patient #1 had a newly described mutation of a splice site predicted to have similar consequence on UBA1 protein (Bourbon et al. Blood 2021). Patient #3 had also somatic mutations in DNMT3A and TP53 (VAF 11% and 2%, respectively), while no other mutation was detected in patients #1 and #2 in our next-generation sequencing panel including 80 genes involved in myeloid malignancies.
3 patients in our MDS/CMML SIAD cohort had relapsing polychondritis, and 2 of them (patients #1 and #2) had UBA1 mutations. The last patient with UBA1 mutation (#3) had giant cell arteritis (GCA) (out of 7 GCA diagnosis in our SIAD cohort). All 3 patients with UBA1 mutation had steroid refractory SIAD and were heavily treated with other immunosuppressive therapies including methotrexate, azathioprine or cytokine targeting agents. Patients #1 and #3 received azacytidine (AZA) for their SIAD, based on our previous experience (Fraison et al. Leukemia Research 2016), with a partial response allowing dose reduction of steroids. At last follow up, none of the 3 patients with UBA1 mutation had MDS progression, and patient #2 died from a stroke 2.4 years after MDS diagnosis.

Conclusion
Our retrospective study represents the first assessment of UBA1 mutation among patients with myeloid malignancies associated with SIAD. The low prevalence of UBA1 mutations in our cohort suggests that other pathophysiological mechanisms may drive inflammation in a majority of MDS/CMML patients with associated SIAD, including mutations in epigenetic regulators TET2/IDH and SRSF2 as we previously proposed.
Keyword(s): Chronic myelomonocytic leukemia, Genetic, Immunology, Myelodysplasia


