
Contributions
Abstract: EP894
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Myelodysplastic syndrome (MDS) and acute myeloid leukemia(AML) are both myeloid neoplasms with obvious heterogeneity. The biological mechanisms of clonal proliferation,abnormal differentiation of hematopoietic stem and progenitor cells(HSPC)are largely unknown. Single-cell RNA sequencing (scRNA-seq) can reveal the specific information of individual cells, which is of great significance for revealing the heterogeneity and exploring the mechanism of MDS and AML.
Aims
To explore the mechanism of clonal proliferation and delayed differentiation of HSPC in patients with MDS and AML at single-cell level.
Methods
5 MDS patients (2 with low-risk MDS[LR-MDS] and 3 with high-risk MDS[HR-MDS]), 2 AML patients were recruited. All the patients have signed the informed consent. Lineage-negative (Lin-) hematopoietic cells from bone marrow(BM) were sorted by immunomagnetic beads,then scRNA-seq and bioinformatics analysis were given. The scRNA-seq data of BM mononuclear cells from 17 healthy donors (HDs) were downloaded from Gene Expression Omnibus (GEO) database, and the data of Lin- cells were selected manually as control. According to the gene-set enrichment analysis, ribosomal genes and mitochondrial genes were selected for further confirmation by Quantitative Real-time polymerase chain reaction (qPCR) in CD34+ cells from BM, and the correlations between gene expression and clinical characteristics were analyzed.
Results
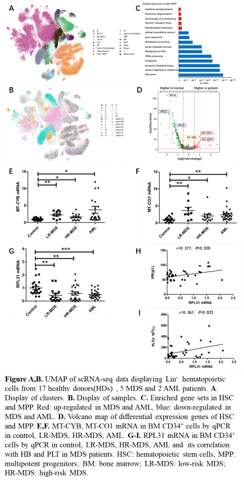
The gene expression patterns in patients with MDS and AML were significantly different from HDs, and they were also significantly heterogeneous among patients. The proportion of hematopoietic stem cells(HSC) and multipotent progenitors(MPP) in MDS and AML was higher than that in HDs, while the proportion of erythroid precursors and lymphoid progenitors was significantly lower than that in HDs(Figure A,B).
Further analysis of HSC and MPP showed that the differential expression genes of MDS compared to HDs were similar to those of AML compared to HDs, so MDS and AML were put together to compare with HDs. Down-regulation of ribosomal genes and the up-regulation of mitochondrial genes were the most significant. Enriched up-regulated gene sets included abnormal mitochondrial metabolism, oxidative respiratory chain defects and neutrophil degranulation in HSC and MPP of MDS and AML patients, while down-regulated gene sets were enriched in ribosome, translation, metabolism of proteins(Figure C,D).
The mRNA expression of ribosomal and mitochondrial genes in BM CD34+ cells by qPCR was consistent with the data of scRNA-seq. The expression of MT-CYB mRNA in LR-MDS(n=10), HR-MDS(n=14), AML(n=26) was significantly higher than that in the control group(patients with iron-deficiency anemia[IDA], n=16)(P<0.05), and the expression of MT-CO1 mRNA got similar results(P<0.05). (Figure E,F). The expression of RPL31 mRNA in LR-MDS(n=14), HR-MDS(n=24), AML(n=25) was lower than that in the control group(patients with IDA, n=22)(P<0.01), and positively correlated with hemoglobin (HB)(r=0.377, P=0.020)and platelet (PLT)(r=0.367, P=0.023). (Figure G-I).
Conclusion
In patients with MDS and AML, dysfunction of ribosome and translation, abnormal mitochondrial metabolism and oxidative respiratory chain in HSPC, may be related to the incidence and development of MDS and AML.
Keyword(s): HSC, MDS/AML
Abstract: EP894
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Myelodysplastic syndrome (MDS) and acute myeloid leukemia(AML) are both myeloid neoplasms with obvious heterogeneity. The biological mechanisms of clonal proliferation,abnormal differentiation of hematopoietic stem and progenitor cells(HSPC)are largely unknown. Single-cell RNA sequencing (scRNA-seq) can reveal the specific information of individual cells, which is of great significance for revealing the heterogeneity and exploring the mechanism of MDS and AML.
Aims
To explore the mechanism of clonal proliferation and delayed differentiation of HSPC in patients with MDS and AML at single-cell level.
Methods
5 MDS patients (2 with low-risk MDS[LR-MDS] and 3 with high-risk MDS[HR-MDS]), 2 AML patients were recruited. All the patients have signed the informed consent. Lineage-negative (Lin-) hematopoietic cells from bone marrow(BM) were sorted by immunomagnetic beads,then scRNA-seq and bioinformatics analysis were given. The scRNA-seq data of BM mononuclear cells from 17 healthy donors (HDs) were downloaded from Gene Expression Omnibus (GEO) database, and the data of Lin- cells were selected manually as control. According to the gene-set enrichment analysis, ribosomal genes and mitochondrial genes were selected for further confirmation by Quantitative Real-time polymerase chain reaction (qPCR) in CD34+ cells from BM, and the correlations between gene expression and clinical characteristics were analyzed.
Results
The gene expression patterns in patients with MDS and AML were significantly different from HDs, and they were also significantly heterogeneous among patients. The proportion of hematopoietic stem cells(HSC) and multipotent progenitors(MPP) in MDS and AML was higher than that in HDs, while the proportion of erythroid precursors and lymphoid progenitors was significantly lower than that in HDs(Figure A,B).
Further analysis of HSC and MPP showed that the differential expression genes of MDS compared to HDs were similar to those of AML compared to HDs, so MDS and AML were put together to compare with HDs. Down-regulation of ribosomal genes and the up-regulation of mitochondrial genes were the most significant. Enriched up-regulated gene sets included abnormal mitochondrial metabolism, oxidative respiratory chain defects and neutrophil degranulation in HSC and MPP of MDS and AML patients, while down-regulated gene sets were enriched in ribosome, translation, metabolism of proteins(Figure C,D).
The mRNA expression of ribosomal and mitochondrial genes in BM CD34+ cells by qPCR was consistent with the data of scRNA-seq. The expression of MT-CYB mRNA in LR-MDS(n=10), HR-MDS(n=14), AML(n=26) was significantly higher than that in the control group(patients with iron-deficiency anemia[IDA], n=16)(P<0.05), and the expression of MT-CO1 mRNA got similar results(P<0.05). (Figure E,F). The expression of RPL31 mRNA in LR-MDS(n=14), HR-MDS(n=24), AML(n=25) was lower than that in the control group(patients with IDA, n=22)(P<0.01), and positively correlated with hemoglobin (HB)(r=0.377, P=0.020)and platelet (PLT)(r=0.367, P=0.023). (Figure G-I).

Conclusion
In patients with MDS and AML, dysfunction of ribosome and translation, abnormal mitochondrial metabolism and oxidative respiratory chain in HSPC, may be related to the incidence and development of MDS and AML.
Keyword(s): HSC, MDS/AML


