
Contributions
Abstract: EP893
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Mutations in cohesin genes have been described as novel mutations occurring in 10-15% of myelodysplastic syndromes (MDS), suggesting that the cohesin complex presents an important pathway in the MDS pathogenesis. However, the clinical impact of cohesin mutations is still undetermined.
Aims
Molecular and clinical characterization of MDS patients harboring cohesin mutations: genetic background, disease phenotype, clinical outcome and prognostic impact of integration of these mutations with conventional risk factors.
Methods
A cohort of 421 patients diagnosed with MDS was analyzed by targeted deep sequencing using an Illumina® custom panel of 117 myeloid-related genes, including cohesin genes: STAG1, STAG2, SMC1A, SMC3, RAD21 and CTCF.
Results
The median age at diagnosis was 74.7 years (range 29.4-92.2); 59% were male. According to the WHO 2017 classification most of the patients had MDS with multilineage dysplasia with or without ring sideroblasts (MDS-RS-MLD and MDS-MLD, 45.5%), followed by MDS with excess blasts (MDS-EB2, 16% and MDS-EB1, 14.6%). Regarding IPSS-R, the majority of patients had very low (27%) and low (44%) risk, with 85% of the series having normal karyotype or clonal alterations of very good or good risk. The median follow-up was 2.3 years (range 0.01-15.6) and during this time 49% of patients died and 30% progressed to acute myeloid leukemia (AML).
The NGS study identified 54 mutations in cohesin genes in 50 patients (11.9%), being most of them loss of function type and STAG2 (36/50) and SMC3 (7/50) the most frequently mutated genes. Regarding the molecular landscape of patients with cohesin mutations: mutated patients presented a significantly higher number of mutations (p<0.0001) and displayed a specific mutational profile with a strong co-occurrence between cohesin and splicing mutations (p<0.0001) and Ras pathway alterations (p=0.001).
Regarding the clinical characterization, mutations in cohesin genes were associated with MDS-EB-1/2 subtypes (p<0.0001), intermediate and high-risk IPSS-R categories (p=0.001) and intermediate cytogenetic risk (p=0.001), especially with trisomy 8 (p<0.0001). Moreover, patients with cohesin mutations displayed a poor prognosis disease phenotype, characterized by lower platelet, neutrophil and leucocyte count in blood (p=0.004, p=0.003 and p=0.037, respectively), a higher number of blasts in bone marrow (p=0.0003) and a higher rate of progression to sAML (p<0.0001).
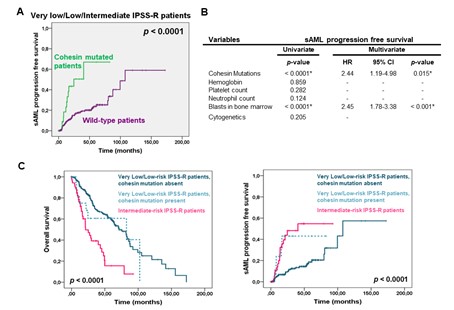
Regarding the influence on clinical outcome, cohesin-mutated patients showed a shorter overall survival (OS: 3.1 vs. 5.2 years, p=0.052) and a shorter time to sAML progression (LFS: 1.4 vs. 8.3 years, p<0.0001). Noteworthy, this adverse impact was observed mainly in very low, low and intermediate-risk IPSS-R patients regarding LFS (multivariate analysis: HR 2.4, 95% CI 1.2-5.0; p=0.015).
To further study the negative impact of cohesin mutations, we assessed their prognostic value on the IPSS-R stratification. Of note, the OS and LFS of very low/low-risk patients who harbored cohesin mutations were statistically significant shorter than those without these mutations, but nearly identical to intermediate-risk patients.
Conclusion
Patients harboring cohesin mutations displayed a specific mutational profile, a poor prognosis disease phenotype and a worse clinical outcome, characterized by a higher rate and a shorter time to sAML progression. In addition, the incorporation of cohesin mutational data into current IPSS-R classification improved the prognostic stratification of very low and low-risk patients.
Keyword(s): Clinical outcome, Mutation analysis, Myelodysplasia, Prognostic factor
Abstract: EP893
Type: E-Poster Presentation
Session title: Myelodysplastic syndromes - Biology & Translational Research
Background
Mutations in cohesin genes have been described as novel mutations occurring in 10-15% of myelodysplastic syndromes (MDS), suggesting that the cohesin complex presents an important pathway in the MDS pathogenesis. However, the clinical impact of cohesin mutations is still undetermined.
Aims
Molecular and clinical characterization of MDS patients harboring cohesin mutations: genetic background, disease phenotype, clinical outcome and prognostic impact of integration of these mutations with conventional risk factors.
Methods
A cohort of 421 patients diagnosed with MDS was analyzed by targeted deep sequencing using an Illumina® custom panel of 117 myeloid-related genes, including cohesin genes: STAG1, STAG2, SMC1A, SMC3, RAD21 and CTCF.
Results
The median age at diagnosis was 74.7 years (range 29.4-92.2); 59% were male. According to the WHO 2017 classification most of the patients had MDS with multilineage dysplasia with or without ring sideroblasts (MDS-RS-MLD and MDS-MLD, 45.5%), followed by MDS with excess blasts (MDS-EB2, 16% and MDS-EB1, 14.6%). Regarding IPSS-R, the majority of patients had very low (27%) and low (44%) risk, with 85% of the series having normal karyotype or clonal alterations of very good or good risk. The median follow-up was 2.3 years (range 0.01-15.6) and during this time 49% of patients died and 30% progressed to acute myeloid leukemia (AML).
The NGS study identified 54 mutations in cohesin genes in 50 patients (11.9%), being most of them loss of function type and STAG2 (36/50) and SMC3 (7/50) the most frequently mutated genes. Regarding the molecular landscape of patients with cohesin mutations: mutated patients presented a significantly higher number of mutations (p<0.0001) and displayed a specific mutational profile with a strong co-occurrence between cohesin and splicing mutations (p<0.0001) and Ras pathway alterations (p=0.001).
Regarding the clinical characterization, mutations in cohesin genes were associated with MDS-EB-1/2 subtypes (p<0.0001), intermediate and high-risk IPSS-R categories (p=0.001) and intermediate cytogenetic risk (p=0.001), especially with trisomy 8 (p<0.0001). Moreover, patients with cohesin mutations displayed a poor prognosis disease phenotype, characterized by lower platelet, neutrophil and leucocyte count in blood (p=0.004, p=0.003 and p=0.037, respectively), a higher number of blasts in bone marrow (p=0.0003) and a higher rate of progression to sAML (p<0.0001).
Regarding the influence on clinical outcome, cohesin-mutated patients showed a shorter overall survival (OS: 3.1 vs. 5.2 years, p=0.052) and a shorter time to sAML progression (LFS: 1.4 vs. 8.3 years, p<0.0001). Noteworthy, this adverse impact was observed mainly in very low, low and intermediate-risk IPSS-R patients regarding LFS (multivariate analysis: HR 2.4, 95% CI 1.2-5.0; p=0.015).
To further study the negative impact of cohesin mutations, we assessed their prognostic value on the IPSS-R stratification. Of note, the OS and LFS of very low/low-risk patients who harbored cohesin mutations were statistically significant shorter than those without these mutations, but nearly identical to intermediate-risk patients.

Conclusion
Patients harboring cohesin mutations displayed a specific mutational profile, a poor prognosis disease phenotype and a worse clinical outcome, characterized by a higher rate and a shorter time to sAML progression. In addition, the incorporation of cohesin mutational data into current IPSS-R classification improved the prognostic stratification of very low and low-risk patients.
Keyword(s): Clinical outcome, Mutation analysis, Myelodysplasia, Prognostic factor


