
Contributions
Abstract: EP858
Type: E-Poster Presentation
Session title: Lymphoma Biology & Translational Research
Background
Mantle cell lymphoma (MCL) is a mature B cell lymphoma with a generally poor prognosis. However, in the past decade a subset of MCL with more indolent clinical course and predominant leukemic disease, termed leukemic non-nodal MCL (nnMCL), has been identified. It is important to confidently diagnose the two MCL subtypes in order to tailor treatment decisions based on their remarkably different clinical course. Current strategies in the differential diagnosis between nnMCL and conventional MCL (cMCL) include SOX11 expression and a gene expression signature (L-MCL16, Clot G et al., Blood 2018), but both methods present limitations which pose difficulties in clinical practice (e.g. low or partial expression of SOX11, L-MCL16 designed for peripheral blood samples). The epigenetic characterization of MCL (Queirós AC et al., Cancer Cell 2016 and Nadeu F et al., Blood 2020) identified two disease subtypes (C1 and C2), which mostly represent cMCL and nnMCL respectively. Furthermore, a SOX11 distal enhancer region was discovered, which is de novo demethylated in SOX11 positive MCL.
Aims
Our study aims to translate the epigenetic subtypes of MCL into a simple assay, which can be easily implemented in clinical practice and universally applied to all types of clinical samples. Secondly, we aim to analyze the impact of the epigenetic disease subtype on clinical outcome in an extended series of cases. Lastly, we aim to characterize the DNA methylation profile of regulatory regions of SOX11 to better understand the relationship between its expression and the C1 and C2 MCL subtypes.
Methods
We identified a 3 CpG signature (Duran-Ferrer M et al., Nature Cancer 2020) that accurately subclassifies MCL into C1 and C2 MCL. To overcome limitations of previous methodologies, we designed pyrosequencing assays that can be applied to any type of clinical sample, including frozen and formalin-fixed paraffin-embedded (FFPE) tissues, as well as peripheral blood samples. Extracted DNA was bisulfite converted, amplified by PCR and pyrosequenced for quantitative methylation analysis. Benchmarking experiments for validation of the assays were performed assessing technical reproducibility and methylation level comparison between different source of samples from the same patient (frozen, FFPE, peripheral blood).
Results
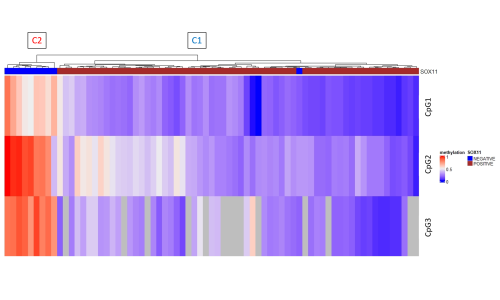
Benchmarking experiments show an excellent technical reproducibility and high correlation between different sample types of the same patient. The exploratory study shows a clear separation in two groups, which correspond to C1 and C2 MCL (Figure). C1 MCL show SOX11 positivity by immunohistochemistry, except for one single cases of pleomorphic MCL. All SOX11 expressing MCL show a consistent demethylation in the SOX11 distal enhancer and promoter region, whereas a majority of SOX11 negative cases have high methylation levels in one of the regulatory regions, indicating that the DNA methylation status of SOX11 regulatory regions might represent a bona fide surrogate of its expression status.
Conclusion
Our study highlights the stability and applicability of a methylation signature to classify MCL subtypes. Analysis of the methylation signature in an extended series of cases is currently ongoing and will allow us to determine the prognostic significance of the epigenetic subtype of MCL. This epigenetic signature applicable to any type of clinical samples may represent a novel and accurate tool for the differential diagnosis of cMCL and nnMCL in clinical routine.
Keyword(s):
Abstract: EP858
Type: E-Poster Presentation
Session title: Lymphoma Biology & Translational Research
Background
Mantle cell lymphoma (MCL) is a mature B cell lymphoma with a generally poor prognosis. However, in the past decade a subset of MCL with more indolent clinical course and predominant leukemic disease, termed leukemic non-nodal MCL (nnMCL), has been identified. It is important to confidently diagnose the two MCL subtypes in order to tailor treatment decisions based on their remarkably different clinical course. Current strategies in the differential diagnosis between nnMCL and conventional MCL (cMCL) include SOX11 expression and a gene expression signature (L-MCL16, Clot G et al., Blood 2018), but both methods present limitations which pose difficulties in clinical practice (e.g. low or partial expression of SOX11, L-MCL16 designed for peripheral blood samples). The epigenetic characterization of MCL (Queirós AC et al., Cancer Cell 2016 and Nadeu F et al., Blood 2020) identified two disease subtypes (C1 and C2), which mostly represent cMCL and nnMCL respectively. Furthermore, a SOX11 distal enhancer region was discovered, which is de novo demethylated in SOX11 positive MCL.
Aims
Our study aims to translate the epigenetic subtypes of MCL into a simple assay, which can be easily implemented in clinical practice and universally applied to all types of clinical samples. Secondly, we aim to analyze the impact of the epigenetic disease subtype on clinical outcome in an extended series of cases. Lastly, we aim to characterize the DNA methylation profile of regulatory regions of SOX11 to better understand the relationship between its expression and the C1 and C2 MCL subtypes.
Methods
We identified a 3 CpG signature (Duran-Ferrer M et al., Nature Cancer 2020) that accurately subclassifies MCL into C1 and C2 MCL. To overcome limitations of previous methodologies, we designed pyrosequencing assays that can be applied to any type of clinical sample, including frozen and formalin-fixed paraffin-embedded (FFPE) tissues, as well as peripheral blood samples. Extracted DNA was bisulfite converted, amplified by PCR and pyrosequenced for quantitative methylation analysis. Benchmarking experiments for validation of the assays were performed assessing technical reproducibility and methylation level comparison between different source of samples from the same patient (frozen, FFPE, peripheral blood).
Results
Benchmarking experiments show an excellent technical reproducibility and high correlation between different sample types of the same patient. The exploratory study shows a clear separation in two groups, which correspond to C1 and C2 MCL (Figure). C1 MCL show SOX11 positivity by immunohistochemistry, except for one single cases of pleomorphic MCL. All SOX11 expressing MCL show a consistent demethylation in the SOX11 distal enhancer and promoter region, whereas a majority of SOX11 negative cases have high methylation levels in one of the regulatory regions, indicating that the DNA methylation status of SOX11 regulatory regions might represent a bona fide surrogate of its expression status.

Conclusion
Our study highlights the stability and applicability of a methylation signature to classify MCL subtypes. Analysis of the methylation signature in an extended series of cases is currently ongoing and will allow us to determine the prognostic significance of the epigenetic subtype of MCL. This epigenetic signature applicable to any type of clinical samples may represent a novel and accurate tool for the differential diagnosis of cMCL and nnMCL in clinical routine.
Keyword(s):


