
Contributions
Abstract: EP755
Type: E-Poster Presentation
Session title: Hematopoiesis, stem cells and microenvironment
Background
Clonal hematopoiesis of indeterminate potential (CHIP) occurs in the peripheral blood of approximately 20% of people > 60 years of age without history of hematologic disorders. CHIP is associated with age related conditions such as cardiovascular diseases and increased all-cause mortality. DNA-methylation at various CpG loci is known to reliably correlate with chronological age and mortality. Different epigenetic clocks have been developed based on these methylation sites and their potential to reflect biological age is a research topic of great interest.
Aims
Aim of the current study was to investigate the relationship between CHIP status, epigenetic age, and epigenetic age acceleration in 382 elderly individuals.
Methods
Epigenetic age was determined using DNA extracted from peripheral blood of elderly individuals (median 74.8; range 55-98 years) without hematologic malignancy and with previously determined CHIP status (Arends et al., Leukemia, 2018). Methylation fractions were determined using methylation-sensitive single nucleotide primer extension. DNA methylation age (DNAm age) was calculated using an adapted version of an 8-CpG clock (Vidal Bralo et al., Front Genet, 2016), implementing a previously established regression model from the Berlin Aging Study II cohort. Age acceleration (AA) was determined as the residual that results from DNAm age regression on chronological age. Intrinsic epigenetic age acceleration (IEAA) was determined likewise with regression on chronological age and blood cell counts.
Results
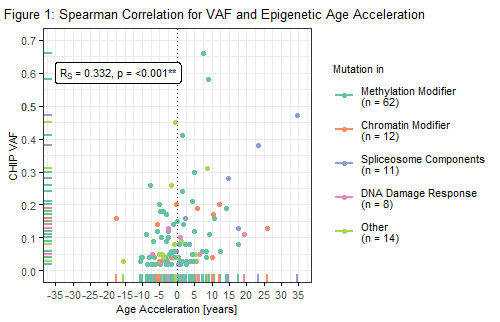
Median DNAm age was 76.15 (SD ± 9.67) years. As determined previously, 107 (28%) of the 382 individuals had evidence of CHIP with a median VAF of 6%. In line with previous publications, sex was significantly associated with AA (female -1.18 years; male 0.96 years; p = .007). When looking at all cases, CHIP was not significantly associated with epigenetic age acceleration (CHIP pos. 1.07 years; CHIP neg. -0.41 years; p = .201). On the contrary, presence of CHIP clones with large VAF (≥ 20%, n = 15) significantly correlated with increased AA when compared to CHIP negative individuals (contrast analysis following ANOVA, p = .022). Mean difference in AA between these groups was 6.04 years. To further analyze the impact of clone size and to account for the continuous character of VAF, a Spearman correlation was performed for AA and VAF of individuals with CHIP (n = 107). Here, we observed a significant, positive correlation between age acceleration and clone size (Rs = .332; p = <.001; Figure 1), which remained significant after adjustment for sex and chronological age (Rs = .326; p = <.001). Comparable results were obtained when correlating IEAA and CHIP clone size (n = 84; Rs = .411; p = <.001 after adjustment for sex and chronological age). When looking at individual functional groups of CHIP genes [methylation modifiers (DNMT3A, TET2), chromatin modifiers (ASXL1, EZH2), spliceosome components (SF3B1, SRSF2, U2AF1, ZRSR2), DNA damage response (TP53, PPM1D), other], no significant correlations could be observed with respect to age acceleration. However, small group sizes preclude final conclusions and warrant further investigation.
Conclusion
In our study investigating the correlation of epigenetic age and CHIP, clone size was significantly associated with epigenetic age acceleration, indicating premature aging in this population. Though cause and consequence relations require further study, these results provide a new perspective on the critical role of CHIP clone size.
Keyword(s): Aging, Epigenetic, Mutation status
Abstract: EP755
Type: E-Poster Presentation
Session title: Hematopoiesis, stem cells and microenvironment
Background
Clonal hematopoiesis of indeterminate potential (CHIP) occurs in the peripheral blood of approximately 20% of people > 60 years of age without history of hematologic disorders. CHIP is associated with age related conditions such as cardiovascular diseases and increased all-cause mortality. DNA-methylation at various CpG loci is known to reliably correlate with chronological age and mortality. Different epigenetic clocks have been developed based on these methylation sites and their potential to reflect biological age is a research topic of great interest.
Aims
Aim of the current study was to investigate the relationship between CHIP status, epigenetic age, and epigenetic age acceleration in 382 elderly individuals.
Methods
Epigenetic age was determined using DNA extracted from peripheral blood of elderly individuals (median 74.8; range 55-98 years) without hematologic malignancy and with previously determined CHIP status (Arends et al., Leukemia, 2018). Methylation fractions were determined using methylation-sensitive single nucleotide primer extension. DNA methylation age (DNAm age) was calculated using an adapted version of an 8-CpG clock (Vidal Bralo et al., Front Genet, 2016), implementing a previously established regression model from the Berlin Aging Study II cohort. Age acceleration (AA) was determined as the residual that results from DNAm age regression on chronological age. Intrinsic epigenetic age acceleration (IEAA) was determined likewise with regression on chronological age and blood cell counts.
Results
Median DNAm age was 76.15 (SD ± 9.67) years. As determined previously, 107 (28%) of the 382 individuals had evidence of CHIP with a median VAF of 6%. In line with previous publications, sex was significantly associated with AA (female -1.18 years; male 0.96 years; p = .007). When looking at all cases, CHIP was not significantly associated with epigenetic age acceleration (CHIP pos. 1.07 years; CHIP neg. -0.41 years; p = .201). On the contrary, presence of CHIP clones with large VAF (≥ 20%, n = 15) significantly correlated with increased AA when compared to CHIP negative individuals (contrast analysis following ANOVA, p = .022). Mean difference in AA between these groups was 6.04 years. To further analyze the impact of clone size and to account for the continuous character of VAF, a Spearman correlation was performed for AA and VAF of individuals with CHIP (n = 107). Here, we observed a significant, positive correlation between age acceleration and clone size (Rs = .332; p = <.001; Figure 1), which remained significant after adjustment for sex and chronological age (Rs = .326; p = <.001). Comparable results were obtained when correlating IEAA and CHIP clone size (n = 84; Rs = .411; p = <.001 after adjustment for sex and chronological age). When looking at individual functional groups of CHIP genes [methylation modifiers (DNMT3A, TET2), chromatin modifiers (ASXL1, EZH2), spliceosome components (SF3B1, SRSF2, U2AF1, ZRSR2), DNA damage response (TP53, PPM1D), other], no significant correlations could be observed with respect to age acceleration. However, small group sizes preclude final conclusions and warrant further investigation.

Conclusion
In our study investigating the correlation of epigenetic age and CHIP, clone size was significantly associated with epigenetic age acceleration, indicating premature aging in this population. Though cause and consequence relations require further study, these results provide a new perspective on the critical role of CHIP clone size.
Keyword(s): Aging, Epigenetic, Mutation status


