
Contributions
Abstract: EP665
Type: E-Poster Presentation
Session title: Chronic myeloid leukemia - Biology & Translational Research
Background
BCR-ABL kinase domain (KD) mutations are the most frequent mechanism of tyrosine kinase inhibitor (TKI) resistance in chronic myeloid leukemia (CML) patients (pts.). Detection of specific mutations influences the choice of the second or subsequent TKI lines. Intensive efforts are focused on characterizing their biological and clinical significance.
Aims
To characterize rare p210 BCR-ABL KD variants revealed in an argentine multicentric study.
Methods
This prospective study screened a cohort of CML TKI resistant pts, from 40 centers, from June 2011 to August 2020. Mutational analysis was performed by direct sequencing of BCR-ABL KD, NCBI sequence reference ID: NM_005157.5 (Branford et al, 2002). Molecular response was defined upon fusion transcript level measured by Real Time quantitative PCR (Gabert et al, 2003) and expressed in international scale as IS%BCR-ABL (Cross et al, 2012).
Results
Mutation screening was performed in 271 CML resistant pts. BCR-ABL DK mutations were detected in 26.2% (71/271) pts: single and compound mutations in 96% (68/71) and 4% (3/71), respectively. The most frequent mutations identified were T315I 35% (25/71) and F317L 11% (8/71). Only 10% (7/71) pts. unveiled rare variants with unknown TKI sensitivity and are described in Table 1. A novel BCR-ABL KD mutation was revealed.
Table 1: Rare BCR-ABL Variants in CML
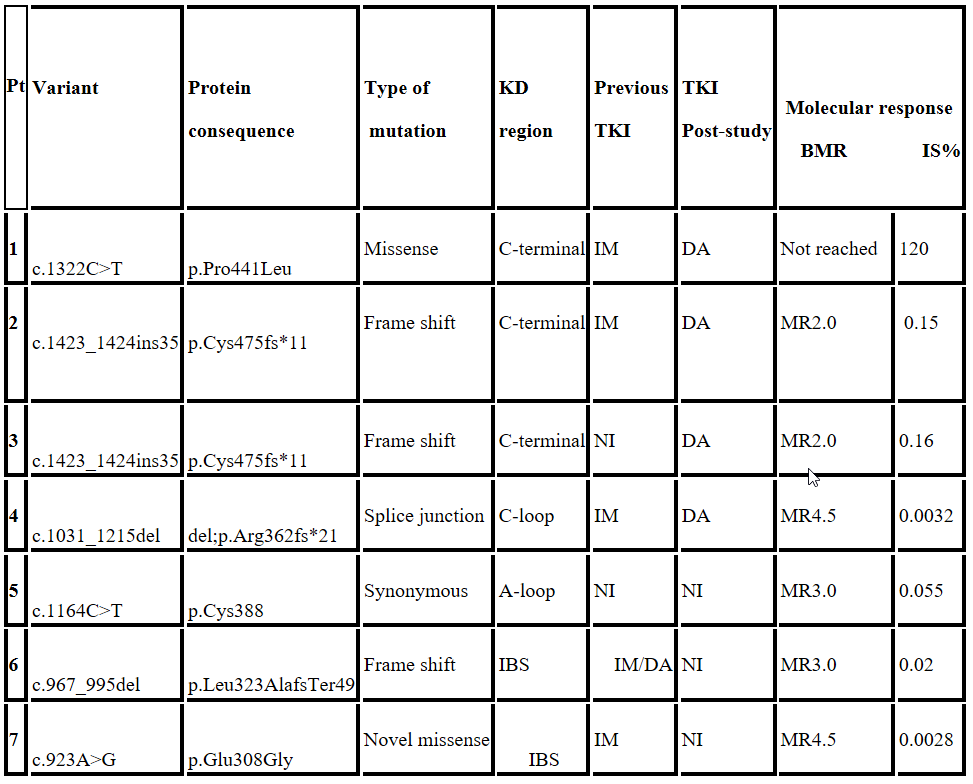
IBS: Imatinib binding site; IM: Imatinib; DA: Dasatinib; NI: Nilotinib; BMR: Best molecular response.
At study entry, all the pts. were in chronic phase, only Pt. 1 evolved to blast crisis independently of the change in TKI. Pts. 2 and 3 exhibited the same 35bp insertion variant, and achieved non optimal responses, while in pts 4, 5, 6 and 7 good MR were obtained: Pts. 4 and 7 achieved deep molecular responses to 2nd generation TKI (MR4.5). A synonymous variant was described in Pt 5 and showed a good molecular evolution despite any TKI treatment changeIBS: Imatinib binding site; IM: Imatinib; DA: Dasatinib; NI: Nilotinib; BMR: Best molecular response.
Conclusion
Detected insertions and deletions variants conducting to truncated proteins, alternative splicing and changes in the Imatinib binding site (IBS) may affect TKI binding to BCR-ABL KD. Variants in the C-terminal region as the missense mutation (c.1322C>T) might confer worse prognosis. Even though the 35bp insertion (c.1423_1424ins35) conferred resistance to Imatinib (IM) and Nilotinib (NI), a sustained suboptimal response was achieved with Dasatinib (DA) therapy. The novel variant (p.Glu308Gly) affects a highly conserved IBS region, probably causing the IM resistance in our patient; however a deep response to NI was achieved. It is important to report single experiences in the treatment and follow up of pts. with uncommon mutations in order to be able to have more data regarding its clinical course. Our study highlights the need for BCR-ABL KD sequence analysis to characterize mutations in therapy resistant CML pts with the aim of guiding targeted treatment.
Keyword(s): BCR-ABL, Chronic myeloid leukemia, Imatinib resistance, Mutation analysis
Abstract: EP665
Type: E-Poster Presentation
Session title: Chronic myeloid leukemia - Biology & Translational Research
Background
BCR-ABL kinase domain (KD) mutations are the most frequent mechanism of tyrosine kinase inhibitor (TKI) resistance in chronic myeloid leukemia (CML) patients (pts.). Detection of specific mutations influences the choice of the second or subsequent TKI lines. Intensive efforts are focused on characterizing their biological and clinical significance.
Aims
To characterize rare p210 BCR-ABL KD variants revealed in an argentine multicentric study.
Methods
This prospective study screened a cohort of CML TKI resistant pts, from 40 centers, from June 2011 to August 2020. Mutational analysis was performed by direct sequencing of BCR-ABL KD, NCBI sequence reference ID: NM_005157.5 (Branford et al, 2002). Molecular response was defined upon fusion transcript level measured by Real Time quantitative PCR (Gabert et al, 2003) and expressed in international scale as IS%BCR-ABL (Cross et al, 2012).
Results
Mutation screening was performed in 271 CML resistant pts. BCR-ABL DK mutations were detected in 26.2% (71/271) pts: single and compound mutations in 96% (68/71) and 4% (3/71), respectively. The most frequent mutations identified were T315I 35% (25/71) and F317L 11% (8/71). Only 10% (7/71) pts. unveiled rare variants with unknown TKI sensitivity and are described in Table 1. A novel BCR-ABL KD mutation was revealed.
Table 1: Rare BCR-ABL Variants in CML

IBS: Imatinib binding site; IM: Imatinib; DA: Dasatinib; NI: Nilotinib; BMR: Best molecular response.
At study entry, all the pts. were in chronic phase, only Pt. 1 evolved to blast crisis independently of the change in TKI. Pts. 2 and 3 exhibited the same 35bp insertion variant, and achieved non optimal responses, while in pts 4, 5, 6 and 7 good MR were obtained: Pts. 4 and 7 achieved deep molecular responses to 2nd generation TKI (MR4.5). A synonymous variant was described in Pt 5 and showed a good molecular evolution despite any TKI treatment changeIBS: Imatinib binding site; IM: Imatinib; DA: Dasatinib; NI: Nilotinib; BMR: Best molecular response.
Conclusion
Detected insertions and deletions variants conducting to truncated proteins, alternative splicing and changes in the Imatinib binding site (IBS) may affect TKI binding to BCR-ABL KD. Variants in the C-terminal region as the missense mutation (c.1322C>T) might confer worse prognosis. Even though the 35bp insertion (c.1423_1424ins35) conferred resistance to Imatinib (IM) and Nilotinib (NI), a sustained suboptimal response was achieved with Dasatinib (DA) therapy. The novel variant (p.Glu308Gly) affects a highly conserved IBS region, probably causing the IM resistance in our patient; however a deep response to NI was achieved. It is important to report single experiences in the treatment and follow up of pts. with uncommon mutations in order to be able to have more data regarding its clinical course. Our study highlights the need for BCR-ABL KD sequence analysis to characterize mutations in therapy resistant CML pts with the aim of guiding targeted treatment.
Keyword(s): BCR-ABL, Chronic myeloid leukemia, Imatinib resistance, Mutation analysis


