
Contributions
Abstract: EP613
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
The human gut microbiome may impact both immune function and carcinogenesis. Previous studies have reported potential associations between gut microbiome constitution and both the incidence and progression of a wide array of cancers. The gut microbiota in patients with chronic lymphocytic leukemia (CLL) remains to be characterized.
Aims
Our research aims to investigate whether gut dysbiosis, i.e. the loss of 'health-promoting' commensal gut microbes and/or the overgrowth of pathogenic bacteria, distinguishes patients with CLL from that of the background population.
Methods
Feces samples were collected, immediately fixated and frozen within 72 h; total genomic DNA was sequenced using Next Generation Sequencing (NGS). Control cohorts were selected from an array of previously published cohorts. Taxonomical profiling was done using an in-house bioinformatics pipeline. Data analysis was performed using standard Compositional Data Analysis (CoDA) methods.
Results
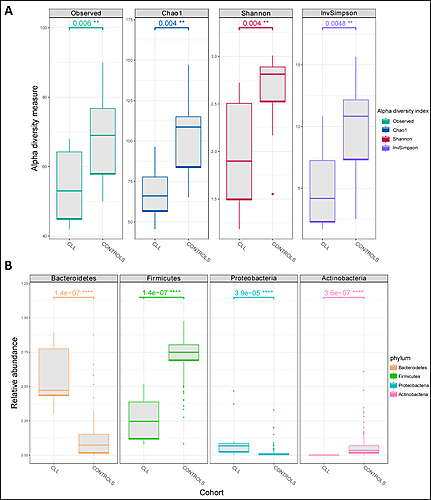
In total 10 CLL patients were included in the study. The control cohort included 124 healthy individuals. We observed that the gut microbiome alpha diversity in bacterial genera was reduced among the CLL patients (Figure 1A). Lower number of observed species (P = 0.006) and a lower diversity (Shannon index, P = 0.004; Inverse Simpson index, P = 0.0048) was observed in the CLL cohort versus healthy controls. Finally, the abundance of Bacteroidetes and Proteobacteria was increased while the abundance of Firmicutes was decreased in the CLL patients compared to healthy controls (Figure 1B).
Conclusion
Our data indicate that the microbiome in CLL patients is less diverse than that of the background population. The gut dysbiosis in CLL includes an increase in Proteobacteria and an increase abundance of other specific bacterial genera. Further investigations are crucial for our understanding of the interplay between the gut microbiome and CLL development. The aim of such studies should be to identify possible interventions modulating the microbiome to change the course of CLL and the adjoined immune dysfunction.
Figure 1. Comparison between the CLL cohort and control cohorts. A) Box plots of Observed number of genera, Chao 1, Shannon and Inverse Simpson indices in CLL cohort and a control cohort. Six controls were randomly sampled from each of the two control cohorts and pooled as CONTROLS. B) Relative abundance of four major bacterial phyla in microbiota in CLL cohort and a control cohort. All samples from each of the two control cohorts were pooled as CONTROLS. The P-values (adjusted for multiple testing with the Benjamini-Hochberg procedure) obtained upon Wilcoxon rank-sum tests are indicated.
Keyword(s): Immune deficiency, Microenvironment, Treatment
Abstract: EP613
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
The human gut microbiome may impact both immune function and carcinogenesis. Previous studies have reported potential associations between gut microbiome constitution and both the incidence and progression of a wide array of cancers. The gut microbiota in patients with chronic lymphocytic leukemia (CLL) remains to be characterized.
Aims
Our research aims to investigate whether gut dysbiosis, i.e. the loss of 'health-promoting' commensal gut microbes and/or the overgrowth of pathogenic bacteria, distinguishes patients with CLL from that of the background population.
Methods
Feces samples were collected, immediately fixated and frozen within 72 h; total genomic DNA was sequenced using Next Generation Sequencing (NGS). Control cohorts were selected from an array of previously published cohorts. Taxonomical profiling was done using an in-house bioinformatics pipeline. Data analysis was performed using standard Compositional Data Analysis (CoDA) methods.
Results
In total 10 CLL patients were included in the study. The control cohort included 124 healthy individuals. We observed that the gut microbiome alpha diversity in bacterial genera was reduced among the CLL patients (Figure 1A). Lower number of observed species (P = 0.006) and a lower diversity (Shannon index, P = 0.004; Inverse Simpson index, P = 0.0048) was observed in the CLL cohort versus healthy controls. Finally, the abundance of Bacteroidetes and Proteobacteria was increased while the abundance of Firmicutes was decreased in the CLL patients compared to healthy controls (Figure 1B).

Conclusion
Our data indicate that the microbiome in CLL patients is less diverse than that of the background population. The gut dysbiosis in CLL includes an increase in Proteobacteria and an increase abundance of other specific bacterial genera. Further investigations are crucial for our understanding of the interplay between the gut microbiome and CLL development. The aim of such studies should be to identify possible interventions modulating the microbiome to change the course of CLL and the adjoined immune dysfunction.
Figure 1. Comparison between the CLL cohort and control cohorts. A) Box plots of Observed number of genera, Chao 1, Shannon and Inverse Simpson indices in CLL cohort and a control cohort. Six controls were randomly sampled from each of the two control cohorts and pooled as CONTROLS. B) Relative abundance of four major bacterial phyla in microbiota in CLL cohort and a control cohort. All samples from each of the two control cohorts were pooled as CONTROLS. The P-values (adjusted for multiple testing with the Benjamini-Hochberg procedure) obtained upon Wilcoxon rank-sum tests are indicated.
Keyword(s): Immune deficiency, Microenvironment, Treatment


