
Contributions
Abstract: EP612
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults in Western countries and remains as an incurable disease. Despite the information provided by genome-wide association studies (GWAS), the genetic component underlying CLL has not been completely unravelled and multiple susceptibility loci remain to be identified. An aberrant expression of autophagy-related genes has been associated with cancer development (1,2) and it has been observed that autophagy shapes anti-tumoral immune responses by acting at multiple levels including the promotion of signals to activate phagocytosis of tumour cells but also signals to induce MHC-class-I and -II presentation (3,4), T-and B-cell activation, development and maintenance (5).
Aims
Considering this background, we investigated whether autophagy-related variants might influence the risk of developing CLL.
Methods
We conducted a population-based case–control study including 1285 Caucasian CLL cases and 1386 healthy controls ascertained through the Consortium for Research in Chronic lymphocytIc LeukemiA (CRuCIAL). Sixteen single nucleotide polymorphisms (SNPs) within autophagy-related loci were selected on the basis of their previous associations with other haematological malignancy (at P<0.01 level) (6). Hardy–Weinberg equilibrium (HWE) was assessed in the control group (P>0.01) and the association between CLL risk and the autophagy-related SNPs was tested using unconditional logistic regression analysis. A P-value of 0.0010 (0.05/16/3 inheritance models) was set to be the multiple testing significance threshold. Functional characterization of the most interesting markers was tested using cytokine quantitative trait loci (cQTL) data after stimulation of peripheral blood mononuclear cells (PBMCs), whole blood or monocyte-derived macrophages (MDMs) from 408 healthy subjects from the Human Functional Genomics Project (HFGP) cohort with LPS, PHA, Pam3Cys, CpG and B. burgdorferi and E. coli, as experimental model for cytokine production capacity. We also analysed the correlation between selected SNPs with data on 91 annotated immune cell populations, 103 serum inflammatory proteins, 7 steroid hormones and autophagy flux.
Results
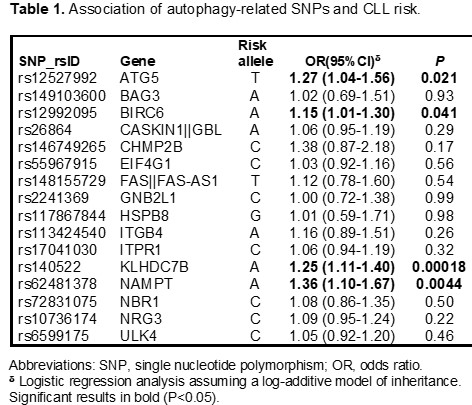
Selected SNPs were in HWE. Logistic regression analysis showed that the ATG5rs12527992, BIRC6rs12992095, KLHDC7Brs140522, and NAMPTrs62481378 SNPs were associated with CLL risk at P<0.05 level. The strongest association was found for the KLHDC7Brs140522 SNP that remained significant after correction for multiple testing. Each copy of the KLHDC7Brs140522A allele increased the risk of CLL by 25% (OR=1.25, 95%CI 1.11-1.40, P=0.0001; Table 1). Furthermore, we found an association for the NAMPTrs62481378 SNP with an increased risk of CLL that remained marginally significant after multiple testing correction. Each copy of the NAMPTrs62481378A allele increased the risk of the disease by 36% (OR=1.36, 95%CI 1.10-1.67, P=0.0044; Table 1). Functional experiments trying to shed some light into the biological mechanisms underlying these associations have been conducted and statistical analyses are currently ongoing.
Conclusion
This study suggests that the KLHDC7B locus is involved in determining the risk of CLL and points to a modest effect of the NAMPT gene. Additional studies are warranted to establish the role of ATG5 and BIRC6 loci in modulating disease risk.
References: [1] Maiuri et al. Cell Death Differ, 2009; [2] Tsuchihara et al. Cancer Lett 2009; [3] Jia et al. J Immunol 2011; [4] Nimmerjahn et al. Eur J Immunol 2003; [5] Arsov et al. J Immunol, 2011; [6] Broderick et al. Nat Genet, 2011.
Keyword(s): Chronic lymphocytic leukemia, Genetic polymorphism, Risk factor
Abstract: EP612
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults in Western countries and remains as an incurable disease. Despite the information provided by genome-wide association studies (GWAS), the genetic component underlying CLL has not been completely unravelled and multiple susceptibility loci remain to be identified. An aberrant expression of autophagy-related genes has been associated with cancer development (1,2) and it has been observed that autophagy shapes anti-tumoral immune responses by acting at multiple levels including the promotion of signals to activate phagocytosis of tumour cells but also signals to induce MHC-class-I and -II presentation (3,4), T-and B-cell activation, development and maintenance (5).
Aims
Considering this background, we investigated whether autophagy-related variants might influence the risk of developing CLL.
Methods
We conducted a population-based case–control study including 1285 Caucasian CLL cases and 1386 healthy controls ascertained through the Consortium for Research in Chronic lymphocytIc LeukemiA (CRuCIAL). Sixteen single nucleotide polymorphisms (SNPs) within autophagy-related loci were selected on the basis of their previous associations with other haematological malignancy (at P<0.01 level) (6). Hardy–Weinberg equilibrium (HWE) was assessed in the control group (P>0.01) and the association between CLL risk and the autophagy-related SNPs was tested using unconditional logistic regression analysis. A P-value of 0.0010 (0.05/16/3 inheritance models) was set to be the multiple testing significance threshold. Functional characterization of the most interesting markers was tested using cytokine quantitative trait loci (cQTL) data after stimulation of peripheral blood mononuclear cells (PBMCs), whole blood or monocyte-derived macrophages (MDMs) from 408 healthy subjects from the Human Functional Genomics Project (HFGP) cohort with LPS, PHA, Pam3Cys, CpG and B. burgdorferi and E. coli, as experimental model for cytokine production capacity. We also analysed the correlation between selected SNPs with data on 91 annotated immune cell populations, 103 serum inflammatory proteins, 7 steroid hormones and autophagy flux.
Results
Selected SNPs were in HWE. Logistic regression analysis showed that the ATG5rs12527992, BIRC6rs12992095, KLHDC7Brs140522, and NAMPTrs62481378 SNPs were associated with CLL risk at P<0.05 level. The strongest association was found for the KLHDC7Brs140522 SNP that remained significant after correction for multiple testing. Each copy of the KLHDC7Brs140522A allele increased the risk of CLL by 25% (OR=1.25, 95%CI 1.11-1.40, P=0.0001; Table 1). Furthermore, we found an association for the NAMPTrs62481378 SNP with an increased risk of CLL that remained marginally significant after multiple testing correction. Each copy of the NAMPTrs62481378A allele increased the risk of the disease by 36% (OR=1.36, 95%CI 1.10-1.67, P=0.0044; Table 1). Functional experiments trying to shed some light into the biological mechanisms underlying these associations have been conducted and statistical analyses are currently ongoing.

Conclusion
This study suggests that the KLHDC7B locus is involved in determining the risk of CLL and points to a modest effect of the NAMPT gene. Additional studies are warranted to establish the role of ATG5 and BIRC6 loci in modulating disease risk.
References: [1] Maiuri et al. Cell Death Differ, 2009; [2] Tsuchihara et al. Cancer Lett 2009; [3] Jia et al. J Immunol 2011; [4] Nimmerjahn et al. Eur J Immunol 2003; [5] Arsov et al. J Immunol, 2011; [6] Broderick et al. Nat Genet, 2011.
Keyword(s): Chronic lymphocytic leukemia, Genetic polymorphism, Risk factor


