
Contributions
Abstract: EP606
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
At a research level, next-generation sequencing (NGS) techniques have provided a large amount of new information, sometimes with clinical implications. In mature B-cell neoplasms (MBCN) the main applications are focused on the identification of somatic variants, while there are limited applications including rearrangements and copy number alterations (CNAs), which are crucial in clinical practice.
Aims
Thus, we designed an NGS capture panel to analyze in a single assay all key genetic aberrations including single nucleotide variants (SNVs), indels, CNAs and chromosomal rearrangements.
Methods
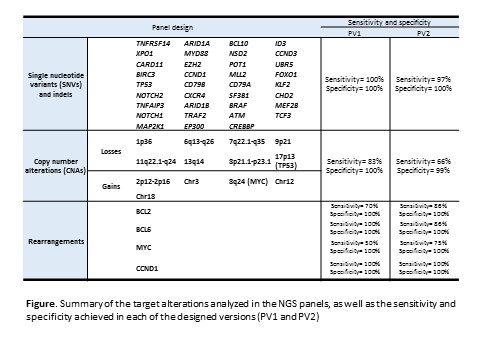
A total of 173 samples from 170 patients diagnosed of different MBCN (55 DLBCL, 43 MZL, 37 CLL, 14 FL, 12 MCL, 5 BL, 4 HCL) were analyzed. DNA was extracted from peripheral blood (n=88), bone marrow (n=7) or other tissues (n=78). Samples were tested in two sequentially designed versions of a custom targeted panel (PV1 and PV2) (Roche Nimblegen) covering 4 rearrangement genes (BCL2, BCL6, MYC and CCND1), 35 frequently mutated genes, and 13 CNA regions (Figure). PV1 also included 9,111 SNPs to assess CNAs genomewide (size=4.6Mb). In PV2 the size was significantly reduced (1.4Mb), and the design for BCL2 and MYC optimized. Sequencing was performed using Illumina NextSeq500. Data were analyzed for SNVs and small indels (MUTECT2), rearrangements (DELLY and Smoove) and CNAs (CopywriteR). Results were validated using different genetic techniques (chromosome banding analysis, FISH, genomic arrays, PCR, Sanger and other NGS panels).
Results
High sensitivity and specificity were achieved (Figure). Regarding mutations, variants with a mean coverage >110x, ≥10 supporting variant reads, and variant allele frequency ≥ 6.4% for PV1 and ≥ 4.2% for PV2 were considered. Besides 107 mutations identified in clinical practice (107/108, 99%), 461 variants affecting the cellular pathways typically described in MBCN were detected, of which 14.1% were classified as pathogenic and 38.7% as likely pathogenic. Regarding rearrangements, optimization of the PV2 design for BCL2 and MYC allowed an improvement in sensitivity. Both panels identified the translocation partners in all patients [including 31% (20/65) in which the partner could not be identified by conventional techniques]. While BCL2 and CCND1 were always rearranged with IGH, 42% of BCL6 rearrangements involved non-IG genes, which were highly variable. MYC was rearranged with IG genes in 72% of cases. In three cases, double rearrangements involving two different translocation partners were detected. Furthermore, a MCL was identified carrying a double rearrangement of MYC with chromosomes 4 and 11, besides CCND1-IGH rearrangement. Regarding CNAs, both panels were highly concordant in terms of size and location of the detected aberrations with those previously defined by genomic arrays. The NGS panels achieved a similar sensitivity than genomic arrays (cut off detection: 20-30% aberrant cells) but the removal of SNPs from PV2 compromised it (Figure).
Conclusion
Our NGS panel allows a more comprehensive evaluation of the genomic landscape of MBCN patients in a single assay by (a) detecting the most important and recurrent mutations, translocations and CNAs with high sensitivity, (b) identifying rearrangement partners not detected by FISH and (c) identifying other clinically relevant genetic alterations not included in clinical guidelines. Although further studies are required to improve its performance, it could potentially be implemented in clinical practice.
Keyword(s): B cell lymphoma, Chronic lymphocytic leukemia, Mutation analysis, Translocation
Abstract: EP606
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
At a research level, next-generation sequencing (NGS) techniques have provided a large amount of new information, sometimes with clinical implications. In mature B-cell neoplasms (MBCN) the main applications are focused on the identification of somatic variants, while there are limited applications including rearrangements and copy number alterations (CNAs), which are crucial in clinical practice.
Aims
Thus, we designed an NGS capture panel to analyze in a single assay all key genetic aberrations including single nucleotide variants (SNVs), indels, CNAs and chromosomal rearrangements.
Methods
A total of 173 samples from 170 patients diagnosed of different MBCN (55 DLBCL, 43 MZL, 37 CLL, 14 FL, 12 MCL, 5 BL, 4 HCL) were analyzed. DNA was extracted from peripheral blood (n=88), bone marrow (n=7) or other tissues (n=78). Samples were tested in two sequentially designed versions of a custom targeted panel (PV1 and PV2) (Roche Nimblegen) covering 4 rearrangement genes (BCL2, BCL6, MYC and CCND1), 35 frequently mutated genes, and 13 CNA regions (Figure). PV1 also included 9,111 SNPs to assess CNAs genomewide (size=4.6Mb). In PV2 the size was significantly reduced (1.4Mb), and the design for BCL2 and MYC optimized. Sequencing was performed using Illumina NextSeq500. Data were analyzed for SNVs and small indels (MUTECT2), rearrangements (DELLY and Smoove) and CNAs (CopywriteR). Results were validated using different genetic techniques (chromosome banding analysis, FISH, genomic arrays, PCR, Sanger and other NGS panels).
Results
High sensitivity and specificity were achieved (Figure). Regarding mutations, variants with a mean coverage >110x, ≥10 supporting variant reads, and variant allele frequency ≥ 6.4% for PV1 and ≥ 4.2% for PV2 were considered. Besides 107 mutations identified in clinical practice (107/108, 99%), 461 variants affecting the cellular pathways typically described in MBCN were detected, of which 14.1% were classified as pathogenic and 38.7% as likely pathogenic. Regarding rearrangements, optimization of the PV2 design for BCL2 and MYC allowed an improvement in sensitivity. Both panels identified the translocation partners in all patients [including 31% (20/65) in which the partner could not be identified by conventional techniques]. While BCL2 and CCND1 were always rearranged with IGH, 42% of BCL6 rearrangements involved non-IG genes, which were highly variable. MYC was rearranged with IG genes in 72% of cases. In three cases, double rearrangements involving two different translocation partners were detected. Furthermore, a MCL was identified carrying a double rearrangement of MYC with chromosomes 4 and 11, besides CCND1-IGH rearrangement. Regarding CNAs, both panels were highly concordant in terms of size and location of the detected aberrations with those previously defined by genomic arrays. The NGS panels achieved a similar sensitivity than genomic arrays (cut off detection: 20-30% aberrant cells) but the removal of SNPs from PV2 compromised it (Figure).

Conclusion
Our NGS panel allows a more comprehensive evaluation of the genomic landscape of MBCN patients in a single assay by (a) detecting the most important and recurrent mutations, translocations and CNAs with high sensitivity, (b) identifying rearrangement partners not detected by FISH and (c) identifying other clinically relevant genetic alterations not included in clinical guidelines. Although further studies are required to improve its performance, it could potentially be implemented in clinical practice.
Keyword(s): B cell lymphoma, Chronic lymphocytic leukemia, Mutation analysis, Translocation


