
Contributions
Abstract: EP603
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Spatial heterogeneity is a hallmark of metastatic solid cancer, while it is less obvious in leukemic B-cell tumors that systemically involve secondary lymphoid organs and circulate in blood. Small lymphocytic lymphoma (SLL) is a model that can inform on whether spatial heterogeneity exists in leukemic B-cell tumors, as it is the sole entity that markedly involves both blood and lymph nodes in all cases. Circulating tumor DNA (ctDNA) recapitulates disease genetics in aggressive lymphomas but its role advantage multiregional sequencing of peripheral blood and lymph node biopsy is unexplored.
Aims
To dissect the genetics of different anatomical compartments in SLL with a multiregional sequencing and liquid biopsy approach.
Methods
Patients with SLL (n=12) were referred to our institution and provided with: i) tumour gDNA extracted from fresh frozen lymph node cells or formalin-fixed paraffin-embedded (FFPE) lymph node biopsies; ii) tumour gDNA extracted from sorted peripheral blood (PB) CD19+/CD5+ cells; iii) ctDNA from plasma; and iv) germline gDNA extracted from CD3+ T-cells and/or granulocytes for comparative purposes. The CAPP-Seq assay was used for the mutational profiling and comprised a panel of 124 genes relevant in B cell malignancies. Copy number variants (CNVs) were identified with the GATK4 CNV workflow.
Results
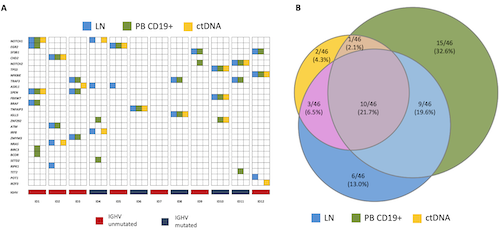
Overall, the analysis of the three SLL compartments analyzed (i.e. lymph node biopsy, circulating PB CD19+ cell compartment, and plasma ctDNA) identified a total of 46 mutations in genes known to be mutated in B cell malignancies. At least one mutation was found in 11 out of 12 (91.7%) patients with a median number of 3 mutations per patient (range 0-6). The most frequently mutated genes were TRAF3 and ASXL1 in 3/12 (25.0%) patients each, followed by NOTCH1, EGR2, and SF3B1 in 2/12 (16.7%) patients each (Fig 1A). By comparing the representation of gene mutations in the different anatomical compartments investigated in SLL, 10/46 (21.7%) mutations were identified in all three compartments, whereas the remaining mutations were differently distributed among the three examined compartments. More precisely, 6/46 (13.0%) mutations were exclusive of the lymph node biopsy, 15/46 (32.6%) were exclusive of the circulating PB CD19+ cell compartment, and only a small fraction of mutations (2/46; 4.3%) were detectable uniquely in the plasma ctDNA (Fig 1B). Interestingly, from a translational perspective, a BIRC3 mutation that may harbor potential predictive value, has been identified only in the circulating PB CD19+ cell compartment. In addition, a bioinformatic algorithm for CNVs analysis has been applied to 8 patients in order to identify potential additional differences between the lymph node biopsy and the circulating PB CD19+ cell compartment. This algorithm showed 100% concordance with FISH karyotype and allowed the detection of at least one CNVs difference in 3/8 patients (37.5%).
Conclusion
These results suggest that the multiregional sequencing of the different anatomical compartments of SLL is essential to gain a comprehensive view of the disease mutational landscape. Consistently, mutational analysis of the SLL lymph node biopsy should be coupled to analysis of the circulating PB CD19+ cell compartment, and eventually with ctDNA analysis. This observation may have clinical relevance when treatment tailoring is based on specific gene mutations used as molecular predictors that might be present in only one specific anatomical compartment of the disease.
Keyword(s): Multiregional sequencing, Small lymphocytic lymphoma
Abstract: EP603
Type: E-Poster Presentation
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Spatial heterogeneity is a hallmark of metastatic solid cancer, while it is less obvious in leukemic B-cell tumors that systemically involve secondary lymphoid organs and circulate in blood. Small lymphocytic lymphoma (SLL) is a model that can inform on whether spatial heterogeneity exists in leukemic B-cell tumors, as it is the sole entity that markedly involves both blood and lymph nodes in all cases. Circulating tumor DNA (ctDNA) recapitulates disease genetics in aggressive lymphomas but its role advantage multiregional sequencing of peripheral blood and lymph node biopsy is unexplored.
Aims
To dissect the genetics of different anatomical compartments in SLL with a multiregional sequencing and liquid biopsy approach.
Methods
Patients with SLL (n=12) were referred to our institution and provided with: i) tumour gDNA extracted from fresh frozen lymph node cells or formalin-fixed paraffin-embedded (FFPE) lymph node biopsies; ii) tumour gDNA extracted from sorted peripheral blood (PB) CD19+/CD5+ cells; iii) ctDNA from plasma; and iv) germline gDNA extracted from CD3+ T-cells and/or granulocytes for comparative purposes. The CAPP-Seq assay was used for the mutational profiling and comprised a panel of 124 genes relevant in B cell malignancies. Copy number variants (CNVs) were identified with the GATK4 CNV workflow.
Results
Overall, the analysis of the three SLL compartments analyzed (i.e. lymph node biopsy, circulating PB CD19+ cell compartment, and plasma ctDNA) identified a total of 46 mutations in genes known to be mutated in B cell malignancies. At least one mutation was found in 11 out of 12 (91.7%) patients with a median number of 3 mutations per patient (range 0-6). The most frequently mutated genes were TRAF3 and ASXL1 in 3/12 (25.0%) patients each, followed by NOTCH1, EGR2, and SF3B1 in 2/12 (16.7%) patients each (Fig 1A). By comparing the representation of gene mutations in the different anatomical compartments investigated in SLL, 10/46 (21.7%) mutations were identified in all three compartments, whereas the remaining mutations were differently distributed among the three examined compartments. More precisely, 6/46 (13.0%) mutations were exclusive of the lymph node biopsy, 15/46 (32.6%) were exclusive of the circulating PB CD19+ cell compartment, and only a small fraction of mutations (2/46; 4.3%) were detectable uniquely in the plasma ctDNA (Fig 1B). Interestingly, from a translational perspective, a BIRC3 mutation that may harbor potential predictive value, has been identified only in the circulating PB CD19+ cell compartment. In addition, a bioinformatic algorithm for CNVs analysis has been applied to 8 patients in order to identify potential additional differences between the lymph node biopsy and the circulating PB CD19+ cell compartment. This algorithm showed 100% concordance with FISH karyotype and allowed the detection of at least one CNVs difference in 3/8 patients (37.5%).

Conclusion
These results suggest that the multiregional sequencing of the different anatomical compartments of SLL is essential to gain a comprehensive view of the disease mutational landscape. Consistently, mutational analysis of the SLL lymph node biopsy should be coupled to analysis of the circulating PB CD19+ cell compartment, and eventually with ctDNA analysis. This observation may have clinical relevance when treatment tailoring is based on specific gene mutations used as molecular predictors that might be present in only one specific anatomical compartment of the disease.
Keyword(s): Multiregional sequencing, Small lymphocytic lymphoma


