
Contributions
Abstract: EP413
Type: E-Poster Presentation
Session title: Acute myeloid leukemia - Biology & Translational Research
Background
Hereditary myeloid malignancies syndromes (HMMSs) are rare but increasingly important clinical entities. These affect ~5-10% of patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). In the 2016 revision, the WHO distinguished them from sporadic myeloid neoplasms, by proposing a specific category: myeloid neoplasms with germline predisposition.
Aims
We prospectively studied the prevalence of HMMSs in a cohort of consecutive patients diagnosed at our institution and explored if there was any association with patients´ medical history.
Methods
So far, we have examined 56 paired somatic/germline samples from 21 MDS and 35 AML consecutive patients (16-80 years). Fifteen (27%) of them were considered secondary neoplasms. The germinal samples included 31 fibroblasts culture from skin biopsy, 22 CD3+ lymphocytes isolated from peripheral blood (PB) and 3 PB in remission. We analyzed them, through targeted Next-Generation sequencing (NGS) using a custom gene panel of >170 genes including all those previously associated to HMMSs. Besides, we conducted a thorough interview with patients to learn about their personal and family history of cancer and signs related to HMMSs.
Results
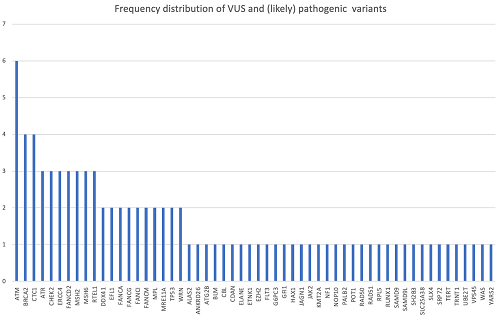
In 10 patients, we did not identify any clinically relevant germinal variant. We found 91 germline variants in 82% (46/56) of patients categorized as variants of uncertain significance (VUS) or (likely) pathogenic according to American College of Medical Genetics and Genomics (ACMG) criteria. Their frequency is shown in figure 1. Mean number of variants per patient was 1.62. In total, 13 (14%) variants were considered as (likely) pathogenic and were present in 12 (21%) patients. Six of them (11% patients) have been associated to HMMSs with an autosomal dominant (AD) inheritance pattern (ANKRD26, CHEK2, ELANE, SAMD9, and TP53 in 2 patients), indicating a potential molecular cause of the disease. Two variants (EZH2, SH2B3) have been related to sporadic myeloid neoplasms but not to a germline origin. Although other heterozygous variants (in genes CTC1, FANCM, SLC25A38, SLX4) have been related to HMMSs, their inheritance pattern is autosomal recessive. Considering those 6 patients with AD (likely) pathogenic variants, mean age was 54 (47-67years). Four of them (67%) presented a personal history of cancer and their myeloid neoplasms were considered secondary to it. In contrast, we noted that only 33% of them exhibited at least one 1st grade relative with cancer and that none of them showed >1 family history of cancer. Patient with CHEK2 and one of those with TP53 mutations presented with typical described phenotype (gynecological cancer history and Li Fraumeni related neoplasms, respectively). However, in the other patients with AD (likely) pathogenic variants, we did not find any sign related to hereditary predisposition syndromes.
Conclusion
In agreement with previous studies, 11% of patients in our series had a potential hereditary myeloid malignancy syndrome, although only 33% of them presented typical phenotypes. Some patients with HMMSs may not initially have suspicious personal or family history due to lack of knowledge about their own or relatives past blood test results, unexplained causes of death in the family, incomplete penetrance, variable expressiveness, or de novo mutations. On the other hand, it is necessary to expand this series to obtain more information on these entities in order to be able to select appropriately those patients who require a germline study and a different clinical management.
Keyword(s): AML, MDS
Abstract: EP413
Type: E-Poster Presentation
Session title: Acute myeloid leukemia - Biology & Translational Research
Background
Hereditary myeloid malignancies syndromes (HMMSs) are rare but increasingly important clinical entities. These affect ~5-10% of patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). In the 2016 revision, the WHO distinguished them from sporadic myeloid neoplasms, by proposing a specific category: myeloid neoplasms with germline predisposition.
Aims
We prospectively studied the prevalence of HMMSs in a cohort of consecutive patients diagnosed at our institution and explored if there was any association with patients´ medical history.
Methods
So far, we have examined 56 paired somatic/germline samples from 21 MDS and 35 AML consecutive patients (16-80 years). Fifteen (27%) of them were considered secondary neoplasms. The germinal samples included 31 fibroblasts culture from skin biopsy, 22 CD3+ lymphocytes isolated from peripheral blood (PB) and 3 PB in remission. We analyzed them, through targeted Next-Generation sequencing (NGS) using a custom gene panel of >170 genes including all those previously associated to HMMSs. Besides, we conducted a thorough interview with patients to learn about their personal and family history of cancer and signs related to HMMSs.
Results
In 10 patients, we did not identify any clinically relevant germinal variant. We found 91 germline variants in 82% (46/56) of patients categorized as variants of uncertain significance (VUS) or (likely) pathogenic according to American College of Medical Genetics and Genomics (ACMG) criteria. Their frequency is shown in figure 1. Mean number of variants per patient was 1.62. In total, 13 (14%) variants were considered as (likely) pathogenic and were present in 12 (21%) patients. Six of them (11% patients) have been associated to HMMSs with an autosomal dominant (AD) inheritance pattern (ANKRD26, CHEK2, ELANE, SAMD9, and TP53 in 2 patients), indicating a potential molecular cause of the disease. Two variants (EZH2, SH2B3) have been related to sporadic myeloid neoplasms but not to a germline origin. Although other heterozygous variants (in genes CTC1, FANCM, SLC25A38, SLX4) have been related to HMMSs, their inheritance pattern is autosomal recessive. Considering those 6 patients with AD (likely) pathogenic variants, mean age was 54 (47-67years). Four of them (67%) presented a personal history of cancer and their myeloid neoplasms were considered secondary to it. In contrast, we noted that only 33% of them exhibited at least one 1st grade relative with cancer and that none of them showed >1 family history of cancer. Patient with CHEK2 and one of those with TP53 mutations presented with typical described phenotype (gynecological cancer history and Li Fraumeni related neoplasms, respectively). However, in the other patients with AD (likely) pathogenic variants, we did not find any sign related to hereditary predisposition syndromes.

Conclusion
In agreement with previous studies, 11% of patients in our series had a potential hereditary myeloid malignancy syndrome, although only 33% of them presented typical phenotypes. Some patients with HMMSs may not initially have suspicious personal or family history due to lack of knowledge about their own or relatives past blood test results, unexplained causes of death in the family, incomplete penetrance, variable expressiveness, or de novo mutations. On the other hand, it is necessary to expand this series to obtain more information on these entities in order to be able to select appropriately those patients who require a germline study and a different clinical management.
Keyword(s): AML, MDS


