
Contributions
Abstract: EP394
Type: E-Poster Presentation
Session title: Acute myeloid leukemia - Biology & Translational Research
Background
In the last years the advent of Next Generation Sequencing (NGS) contributed to the identification of new somatic mutations involved in the leukemogenesis process, allowing a revised stratification of the biological risk for acute myeloid leukemia (AML) and providing a new tool for minimal residual disease evaluation.
Aims
We evaluated the impact between NGS profile and post transplant outcome, with particular interent on relapse occurrence.
Methods
We enrolled 64 patients with AML submitted to allogeneic stem cell transplantation (HSCT) in our department between November 2015 and September 2020. NGS panel performed at diagnosis included 18 genes: CBL, SF3B1, U2AF1, ZRSR2, KRAS, ASXL1, CEBPA, KIT, DNMT3A, EZH2, IDH1, IDH2, RUNX1, SRSF2, TET2, TP53, WT1, NRAS.
Results
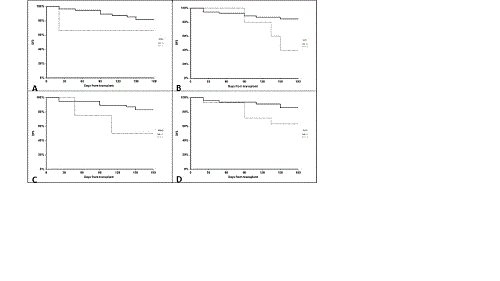
Mutation distribution was as follows: CBL (n=1), SF3B1 (n=1), U2AF1 (n=1), ZRSR2 (n=1), KRAS (n=1), ASXL1 (n=9), CEBPA (n=9), KIT (n=15), DNMT3A (n=22), EZH2 (n=17), IDH1 (n=10), IDH2 (n=10), RUNX1 (n=6), SRSF2 (n=2), TET2 (n=30), TP53 (n=3), WT1 (n=6), NRAS (n=4). Median age was 55.5 years (17-73). Risk group according to European Leukemia Net was as follows: favourable (n=8), intermediate (n=42), adverse (n=14). Thirty-nine patients achieved a first complete remission before transplant, 8 were in second remission, 14 had relapsed disease and 3 received HSCT upfront. A significant difference in terms of DFS was found accordingly to some mutations. Patients mutated for TP53 (n=3) showed a 6 months DFS of 66.7% as compared to 81.4% in non mutated (n=61) (p=0.02, Fig.1A). Mutated WT1 patients (n=6) had a 6 months DFS of 40% as compared to 84.8% in non mutated (n=58) (p=0.047, Fig.1B). Six-months DFS was of 63.5% and 83% in patients with (n=4) and without (n=60) NRAS mutation, respectively (p=0.005, Fig1C). Accordingly to FLT3 mutation, 6 months DFS was of 63.5% in mutated patients (n=14) as compared to 83% for non mutated (n=49) (p=0.048, Fig1D). Multivariate analysis for DFS confirmed TP53 (HR 12.96, 95% CI 4.16-39.61, p=0.0009), WT1 (HR 5.03, 95% CI 1.08-23.37, p=0.04), FLT3 (HR 4.63, 95% CI 1.22-17.50, p=0.02) and NRAS (HR 8.17, 95% CI 4.42-39.62, p=0.0004) together with absence of complete remission at the time of transplant (HR 7.89, 95% CI 2.23-27.92, p=0.001) as independent factors for relapse after transplant. Moreover, we found a detrimental impact of triple mutation FLT3+NPM1+DNMT3A on DFS. Patients with any of these mutations (n=30) had 6 months DFS of 85.2% as compared to 89.8% for patients with one or two mutations (n=23) and 50% in patients triple mutated (n=8) (p=0.04). We then evaluated if a difference might be in young (<60 years) and old patients in terms of mutations influence. TP53, WT1, FLT3 and triple mutation DNMT3A, FLT3 and NPM1 were confirmed in young whereas only NRAS was confirmed in old patients.
Figure 1: 6-months DFS according to TP53 (A), WT1 (B) NRAS (C) and FLT3 (D) mutations.
Conclusion
The presence of some mutations like as WT1, TP53, NRAS, FLT3 or concomitant mutated status for FLT3, NPM1 and DNMT3A worsened post transplant outcome in terms of disease free survival, independently from transplant features. Certainly, these finding need to be confirmed in a larger study population.
Keyword(s): AML, HSCT, Molecular markers
Abstract: EP394
Type: E-Poster Presentation
Session title: Acute myeloid leukemia - Biology & Translational Research
Background
In the last years the advent of Next Generation Sequencing (NGS) contributed to the identification of new somatic mutations involved in the leukemogenesis process, allowing a revised stratification of the biological risk for acute myeloid leukemia (AML) and providing a new tool for minimal residual disease evaluation.
Aims
We evaluated the impact between NGS profile and post transplant outcome, with particular interent on relapse occurrence.
Methods
We enrolled 64 patients with AML submitted to allogeneic stem cell transplantation (HSCT) in our department between November 2015 and September 2020. NGS panel performed at diagnosis included 18 genes: CBL, SF3B1, U2AF1, ZRSR2, KRAS, ASXL1, CEBPA, KIT, DNMT3A, EZH2, IDH1, IDH2, RUNX1, SRSF2, TET2, TP53, WT1, NRAS.
Results
Mutation distribution was as follows: CBL (n=1), SF3B1 (n=1), U2AF1 (n=1), ZRSR2 (n=1), KRAS (n=1), ASXL1 (n=9), CEBPA (n=9), KIT (n=15), DNMT3A (n=22), EZH2 (n=17), IDH1 (n=10), IDH2 (n=10), RUNX1 (n=6), SRSF2 (n=2), TET2 (n=30), TP53 (n=3), WT1 (n=6), NRAS (n=4). Median age was 55.5 years (17-73). Risk group according to European Leukemia Net was as follows: favourable (n=8), intermediate (n=42), adverse (n=14). Thirty-nine patients achieved a first complete remission before transplant, 8 were in second remission, 14 had relapsed disease and 3 received HSCT upfront. A significant difference in terms of DFS was found accordingly to some mutations. Patients mutated for TP53 (n=3) showed a 6 months DFS of 66.7% as compared to 81.4% in non mutated (n=61) (p=0.02, Fig.1A). Mutated WT1 patients (n=6) had a 6 months DFS of 40% as compared to 84.8% in non mutated (n=58) (p=0.047, Fig.1B). Six-months DFS was of 63.5% and 83% in patients with (n=4) and without (n=60) NRAS mutation, respectively (p=0.005, Fig1C). Accordingly to FLT3 mutation, 6 months DFS was of 63.5% in mutated patients (n=14) as compared to 83% for non mutated (n=49) (p=0.048, Fig1D). Multivariate analysis for DFS confirmed TP53 (HR 12.96, 95% CI 4.16-39.61, p=0.0009), WT1 (HR 5.03, 95% CI 1.08-23.37, p=0.04), FLT3 (HR 4.63, 95% CI 1.22-17.50, p=0.02) and NRAS (HR 8.17, 95% CI 4.42-39.62, p=0.0004) together with absence of complete remission at the time of transplant (HR 7.89, 95% CI 2.23-27.92, p=0.001) as independent factors for relapse after transplant. Moreover, we found a detrimental impact of triple mutation FLT3+NPM1+DNMT3A on DFS. Patients with any of these mutations (n=30) had 6 months DFS of 85.2% as compared to 89.8% for patients with one or two mutations (n=23) and 50% in patients triple mutated (n=8) (p=0.04). We then evaluated if a difference might be in young (<60 years) and old patients in terms of mutations influence. TP53, WT1, FLT3 and triple mutation DNMT3A, FLT3 and NPM1 were confirmed in young whereas only NRAS was confirmed in old patients.
Figure 1: 6-months DFS according to TP53 (A), WT1 (B) NRAS (C) and FLT3 (D) mutations.

Conclusion
The presence of some mutations like as WT1, TP53, NRAS, FLT3 or concomitant mutated status for FLT3, NPM1 and DNMT3A worsened post transplant outcome in terms of disease free survival, independently from transplant features. Certainly, these finding need to be confirmed in a larger study population.
Keyword(s): AML, HSCT, Molecular markers


