
Contributions
Abstract: EP340
Type: E-Poster Presentation
Session title: Acute lymphoblastic leukemia - Biology & Translational Research
Background
Prognosis of acute lymphoblastic leukemia (ALL) in adults is inferior than children. Hence, ALL is still a challenging disease to be cured in adult population. Aberrant genetic alterations has been observed previously in ALL, while the patterns of differential gene alteration have not much been comprehensively determined in the adult and pediatric ALL on a genome-wide scale.
Aims
This study attempted to investigate the biological differences in genomic profiling between the adults and children with ALL, and the relationship between the genomic heterogeneity and prognosis.
Methods
39 adults and 54 children patients with ALL in our institution were studied. All patients were de novo and treated with standard chemotherapy regimens. Clinical data were collected on all patients. Exome sequencing and anaiysis was performed. The genomic mutation was calculated. The driver genes of the mutated genes were selected from the COSMIC platform. The overlapped gene was subjected to Gene ontology and pathway enrichment analyses and mapped the PPI network.
Results
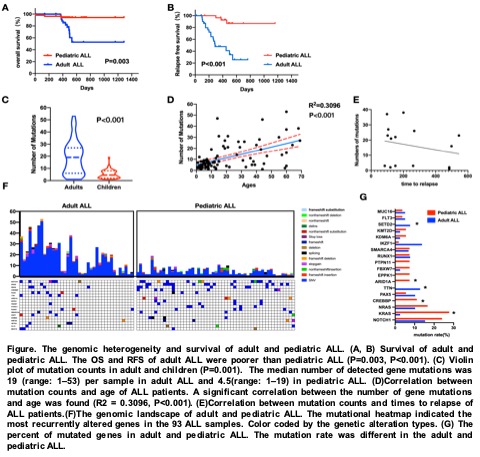
The results showed the similar common mutation types but the increased prevalence of genetic alterations in adult ALL. The median number of detected gene mutations was 19 (range: 1–53) per sample in adult ALL and 4.5 (range: 1–19) in pediatric ALL(P<0.001). A significant correlation between the increased number of gene mutations and age was found (R2 = 0.3096, P<0.001). 67 and 53 driver genes were identified in adult and pediatric ALL samples, respectively. The SETD2 and TTN mutations were significantly enriched in adult ALL. While the KRAS, ARID1A and CREBBP mutations were significantly enriched in pediatric ALL. (P<0.05). 67 and 53 driver genes were identified in adult and pediatric ALL samples, respectively. Using integrated genomic analysis, we identified 4 functional pathways recurrently mutated in adult ALL: transcriptional regulation, VEGF signaling pathway, T cell receptor signaling pathway, and Fc epsilon RI signaling pathway(P<0.05), 5 in pediatric ALL: Transcriptional regulation, PI3K-AKT-mTOR signaling, JAK-STAT signaling, NOTCH1 signaling, FoxO signaling pathway(P<0.05). Using cytoHubba analysis, among the 53 nodes, NRAS, IDH1, WT1, FLT3, NF1, DNMT3A, CEBPA, PTPN11, TP53, ASXL1, the ten central node genes with degree >10, were considered as hub driver genes in the PPI network of adult ALL. While as to the pediatric ALL, MYC, EZH2, CDKN2A, CREBBP, SMARCA4, ATRX, NF1, EP300, NOTCH1, BRCA1, the ten central node genes with degree >10, were considered as hub driver genes in the PPI network.The incidence of relapse was 33.3% and 7.7% in the adult and pediatric ALL, respectively(P=0.003). The overall survival(OS) and relapse free survival (RFS) of adult ALL were poorer than pediatric ALL(P=0.003, P<0.001, respectively).
Conclusion
This genomic landscape enhanced the understanding of the biological differences of ALL between the two populations and provided a clue for novel therapeutic approaches.
Keyword(s): Acute lymphoblastic leukemia, Adult
Abstract: EP340
Type: E-Poster Presentation
Session title: Acute lymphoblastic leukemia - Biology & Translational Research
Background
Prognosis of acute lymphoblastic leukemia (ALL) in adults is inferior than children. Hence, ALL is still a challenging disease to be cured in adult population. Aberrant genetic alterations has been observed previously in ALL, while the patterns of differential gene alteration have not much been comprehensively determined in the adult and pediatric ALL on a genome-wide scale.
Aims
This study attempted to investigate the biological differences in genomic profiling between the adults and children with ALL, and the relationship between the genomic heterogeneity and prognosis.
Methods
39 adults and 54 children patients with ALL in our institution were studied. All patients were de novo and treated with standard chemotherapy regimens. Clinical data were collected on all patients. Exome sequencing and anaiysis was performed. The genomic mutation was calculated. The driver genes of the mutated genes were selected from the COSMIC platform. The overlapped gene was subjected to Gene ontology and pathway enrichment analyses and mapped the PPI network.
Results
The results showed the similar common mutation types but the increased prevalence of genetic alterations in adult ALL. The median number of detected gene mutations was 19 (range: 1–53) per sample in adult ALL and 4.5 (range: 1–19) in pediatric ALL(P<0.001). A significant correlation between the increased number of gene mutations and age was found (R2 = 0.3096, P<0.001). 67 and 53 driver genes were identified in adult and pediatric ALL samples, respectively. The SETD2 and TTN mutations were significantly enriched in adult ALL. While the KRAS, ARID1A and CREBBP mutations were significantly enriched in pediatric ALL. (P<0.05). 67 and 53 driver genes were identified in adult and pediatric ALL samples, respectively. Using integrated genomic analysis, we identified 4 functional pathways recurrently mutated in adult ALL: transcriptional regulation, VEGF signaling pathway, T cell receptor signaling pathway, and Fc epsilon RI signaling pathway(P<0.05), 5 in pediatric ALL: Transcriptional regulation, PI3K-AKT-mTOR signaling, JAK-STAT signaling, NOTCH1 signaling, FoxO signaling pathway(P<0.05). Using cytoHubba analysis, among the 53 nodes, NRAS, IDH1, WT1, FLT3, NF1, DNMT3A, CEBPA, PTPN11, TP53, ASXL1, the ten central node genes with degree >10, were considered as hub driver genes in the PPI network of adult ALL. While as to the pediatric ALL, MYC, EZH2, CDKN2A, CREBBP, SMARCA4, ATRX, NF1, EP300, NOTCH1, BRCA1, the ten central node genes with degree >10, were considered as hub driver genes in the PPI network.The incidence of relapse was 33.3% and 7.7% in the adult and pediatric ALL, respectively(P=0.003). The overall survival(OS) and relapse free survival (RFS) of adult ALL were poorer than pediatric ALL(P=0.003, P<0.001, respectively).

Conclusion
This genomic landscape enhanced the understanding of the biological differences of ALL between the two populations and provided a clue for novel therapeutic approaches.
Keyword(s): Acute lymphoblastic leukemia, Adult


