
Contributions
Abstract: EP1131
Type: E-Poster Presentation
Session title: Platelet disorders
Background
Inherited platelet disorders (IPD) are a heterogeneous group of rare diseases, whose functional and molecular diagnosis has traditionally been complex. Although, high-throughput sequencing (HTS) has allowed us to increase the number of patients with an accurate genetic diagnosis, there are many specific and/or syndromic IPD patients with unknown underlying molecular variations. So, it is probably challenging by the limitation to identify large and complex deletions or amplifications of DNA segments between 50bp and 1kb, called Copy Number Variations (CNV). Comparative Genetic Hybridization array (aCGH) or Multiplex Ligation-dependent Probe Amplification (MLPA) has become the gold standard for constitutional genetic diagnosis of CNV and big structural chromosomal variants, translocations and aneuploidy. The application of HTS to detect CNV is being studied in IPD.
Aims
To identify CNVs using a custom HTS panel in patients with specific and/or syndromic IPDs.
Methods
Custom HTS panel was designed using the SureDesign Studio (Agilent Technologies, Santa Clara, CA, USA) with 57,070 probes, including coding regions for 108 genes with relevance in inherited bleeding disorders, which also allowed to detect CNVs. Regions of interest were enriched for each library following the SureSelectQXT-Target-Enrichment protocol (Agilent), and finally were sequenced on an Illumina NextSeq platform, with a read length of 2x150 nucleotides. Data were processed using an in-house pipeline. CNV analysis were made using depth-of-coverage method, which assumes a linear correlation between the coverage depth and the number of copies. Lower reading depth than expected represented losses or deletions, while higher depth represented gains or amplifications.
Results
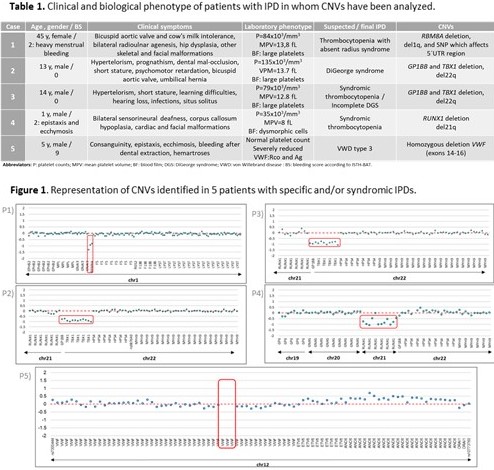
CNVs were detected in 5 patients whose, clinical and laboratory phenotypes were included in Table 1. For CNV analysis an average of 1,8x106 reads per sample was obtained with a minimum mean coverage of 300X that allowed detecting deletions in the following genes: RBM8A, GP1BB, TBX1, RUNX1 and VWF. In patient 1 (P1), thrombocytopenia-absent radius syndrome was previously diagnosed and HTS allowed us to identify a low frequency SNP which affects the 5’UTR region of RBM8A (c.-21 G>A; rs139428292) and a deletion on chromosome 1q. The latter was validated with aCGH showing a microdeletion at chromosome band 1q21.1. In P2, Di George syndrome (DGS) was diagnosed and deletion of GP1BB and TBX1 confirmed a del22q. Additionally, in P3, a syndromic thrombocytopenia (table 1) was suspected and a deletion of GP1BB and TBX1 was detected related to incomplete DGS. In these 2 patients, deletions were validated with MLPA and aCGH, respectively. A deletion on chromosome 21q which involved RUNX1 was identified in P4 with syndromic thrombocytopenia. Finally, a homozygous deletion of 3 exons (14-16) of VWF, was detected in P5 (von Willebrand disease type 3) (Figure 1).
Conclusion
Our custom HTS panel has allowed us not only identify single-nucleotide variants but also CNVs. CNV analysis is particularly useful in IPDs patients with specific phenotype with unknow underlying genetic variants and/or a syndromic thrombocytopenias.
Keyword(s): Inherited platelet disorders
Abstract: EP1131
Type: E-Poster Presentation
Session title: Platelet disorders
Background
Inherited platelet disorders (IPD) are a heterogeneous group of rare diseases, whose functional and molecular diagnosis has traditionally been complex. Although, high-throughput sequencing (HTS) has allowed us to increase the number of patients with an accurate genetic diagnosis, there are many specific and/or syndromic IPD patients with unknown underlying molecular variations. So, it is probably challenging by the limitation to identify large and complex deletions or amplifications of DNA segments between 50bp and 1kb, called Copy Number Variations (CNV). Comparative Genetic Hybridization array (aCGH) or Multiplex Ligation-dependent Probe Amplification (MLPA) has become the gold standard for constitutional genetic diagnosis of CNV and big structural chromosomal variants, translocations and aneuploidy. The application of HTS to detect CNV is being studied in IPD.
Aims
To identify CNVs using a custom HTS panel in patients with specific and/or syndromic IPDs.
Methods
Custom HTS panel was designed using the SureDesign Studio (Agilent Technologies, Santa Clara, CA, USA) with 57,070 probes, including coding regions for 108 genes with relevance in inherited bleeding disorders, which also allowed to detect CNVs. Regions of interest were enriched for each library following the SureSelectQXT-Target-Enrichment protocol (Agilent), and finally were sequenced on an Illumina NextSeq platform, with a read length of 2x150 nucleotides. Data were processed using an in-house pipeline. CNV analysis were made using depth-of-coverage method, which assumes a linear correlation between the coverage depth and the number of copies. Lower reading depth than expected represented losses or deletions, while higher depth represented gains or amplifications.
Results
CNVs were detected in 5 patients whose, clinical and laboratory phenotypes were included in Table 1. For CNV analysis an average of 1,8x106 reads per sample was obtained with a minimum mean coverage of 300X that allowed detecting deletions in the following genes: RBM8A, GP1BB, TBX1, RUNX1 and VWF. In patient 1 (P1), thrombocytopenia-absent radius syndrome was previously diagnosed and HTS allowed us to identify a low frequency SNP which affects the 5’UTR region of RBM8A (c.-21 G>A; rs139428292) and a deletion on chromosome 1q. The latter was validated with aCGH showing a microdeletion at chromosome band 1q21.1. In P2, Di George syndrome (DGS) was diagnosed and deletion of GP1BB and TBX1 confirmed a del22q. Additionally, in P3, a syndromic thrombocytopenia (table 1) was suspected and a deletion of GP1BB and TBX1 was detected related to incomplete DGS. In these 2 patients, deletions were validated with MLPA and aCGH, respectively. A deletion on chromosome 21q which involved RUNX1 was identified in P4 with syndromic thrombocytopenia. Finally, a homozygous deletion of 3 exons (14-16) of VWF, was detected in P5 (von Willebrand disease type 3) (Figure 1).

Conclusion
Our custom HTS panel has allowed us not only identify single-nucleotide variants but also CNVs. CNV analysis is particularly useful in IPDs patients with specific phenotype with unknow underlying genetic variants and/or a syndromic thrombocytopenias.
Keyword(s): Inherited platelet disorders


