
Contributions
Abstract: S297
Type: Oral Presentation
Session title: ITP: from bench to bedside
Background
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disease characterized by isolated thrombocytopenia. A growing body of emerging evidence indicates that abnormalities during any stage of thrombopoiesis and megakaryocytopoiesis can influence platelet production.
Aims
Our study concentrated on the hematopoietic stem and progenitor cells (HSPCs) in ITP patients from the perspective of single cell transcriptome.
Methods
CD34+ HSPCs were isolated from BM of four newly diagnosed ITP patients and four healthy adults as controls by fluorescence-activated cell sorter (FACS), and Single-cell RNA sequencing (scRNA-seq) data was collected using the recommended protocol for the 3’ scRNA-seq 10X genomics platform.
Results
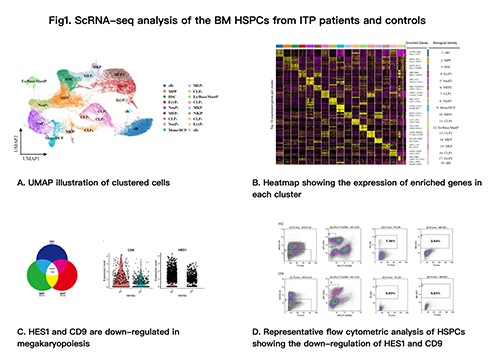
We perform scRNA-seq for 69,178 single CD34+ HSPCs, and detected over 3000 expressed genes per cell on average. Uniform manifold approximation and projection (UMAP), a method for nonlinear dimensionality reduction, showed a visualized image of the cells (Fig1. A). Heatmap showed the scaled expression of top 10 differentially expressed genes in each cluster (Fig1. B). Cell clusters expressed established markers of hematopoietic populations, revealed diverse hematopoietic cell types and implied differentiation trajectories consistent with current views of hematopoiesis. We identified an unambiguous feature of HSPCs in ITP patients with the down-regulation of HES1 and CD9 (Fig1. C). And the results were verified by flow cytometry (Fig1. D). Furthermore, through cell culture, we found that lin-CD45ra-CD34+CD9+ cells produced fewer mature megakaryocytes in ITP patients than healthy controls. Our study suggested that HES1 and CD9 were involved in the pathogenesis of ITP.
Conclusion
Using scRNA-seq, we revealed a hierarchically-structured transcriptional landscape of hematopoietic differentiation of BM CD34+ HSPCs. We observed a significantly descreased expression of HES1 and CD9 in newly diagnosed ITP patients, which might relate with the generation of abnormal MKs and be a biomarker potentially using in diagnosis.
Keyword(s): Hematopoietic stem and progenitor cells, ITP
Abstract: S297
Type: Oral Presentation
Session title: ITP: from bench to bedside
Background
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disease characterized by isolated thrombocytopenia. A growing body of emerging evidence indicates that abnormalities during any stage of thrombopoiesis and megakaryocytopoiesis can influence platelet production.
Aims
Our study concentrated on the hematopoietic stem and progenitor cells (HSPCs) in ITP patients from the perspective of single cell transcriptome.
Methods
CD34+ HSPCs were isolated from BM of four newly diagnosed ITP patients and four healthy adults as controls by fluorescence-activated cell sorter (FACS), and Single-cell RNA sequencing (scRNA-seq) data was collected using the recommended protocol for the 3’ scRNA-seq 10X genomics platform.
Results
We perform scRNA-seq for 69,178 single CD34+ HSPCs, and detected over 3000 expressed genes per cell on average. Uniform manifold approximation and projection (UMAP), a method for nonlinear dimensionality reduction, showed a visualized image of the cells (Fig1. A). Heatmap showed the scaled expression of top 10 differentially expressed genes in each cluster (Fig1. B). Cell clusters expressed established markers of hematopoietic populations, revealed diverse hematopoietic cell types and implied differentiation trajectories consistent with current views of hematopoiesis. We identified an unambiguous feature of HSPCs in ITP patients with the down-regulation of HES1 and CD9 (Fig1. C). And the results were verified by flow cytometry (Fig1. D). Furthermore, through cell culture, we found that lin-CD45ra-CD34+CD9+ cells produced fewer mature megakaryocytes in ITP patients than healthy controls. Our study suggested that HES1 and CD9 were involved in the pathogenesis of ITP.

Conclusion
Using scRNA-seq, we revealed a hierarchically-structured transcriptional landscape of hematopoietic differentiation of BM CD34+ HSPCs. We observed a significantly descreased expression of HES1 and CD9 in newly diagnosed ITP patients, which might relate with the generation of abnormal MKs and be a biomarker potentially using in diagnosis.
Keyword(s): Hematopoietic stem and progenitor cells, ITP


