
Contributions
Abstract: S232
Type: Oral Presentation
Session title: Stem cell transplantation - Preclinical/experimental
Background
TP53 mutations in patients (pts) with acute myeloid leukaemia (AML) has been associated with an unfavourable outcome including after allogeneic stem cell transplantation (allo-SCT). This has led some to question the benefit of allo-SCT for TP53 mutant AML, although it has not been studied in a large patient cohort.
Aims
To identify factors which influence transplant outcome in pts with mutant TP53 AML.
Methods
We identified pts with AML in first complete remission, and with either intermediate or adverse risk cytogenetics, whose TP53 mutation status was reported to the EBMT registry. All ages, conditioning intensity or donor source were included.
Results
We identified 179 pts with AML with TP53 mutations and compared their characteristics and outcomes to 601 pts with AML without TP53 mutations. Pts were transplanted between 2015-2019. Chromosomal losses in chromosomes 17p (31% vs. 3%), 5q (49% vs. 5%), and 7q (34% vs. 8%) as well as complex (65% vs. 12%) and monosomal karyotypes (41% vs. 8%) were significantly increased in the cohort of pts with mutated TP53, in contrast to those with unmutated TP53.
The overall survival (OS) at 2 years was significantly impaired in pts with mutated TP53 as compared to the cohort without (mutated TP53, 35.1% [95% CI: 26.7-43.7]; without TP53 mutations, 64% [95% CI: 59.1-68.4], p= 0.001). This was a result of an increased cumulative incidence of relapse (CIR) at 2 years in pts with mutated TP53 as compared to the cohort without mutated TP53 (mutated TP53, 55% [95% CI: 45.2-63.8]; without TP53 mutations, 25.2% [95%CI: 21.2-29.3], p= 0.001). The adverse prognostic impact of TP53 mutations on OS (HR=1.5, 95%CI 1.1-2.1) and CIR (HR=1.4, 95%CI: 1.2-1.9) was maintained in a multivariate model that accounted for the adverse prognostic value of other known cytogenetic, and transplant, variables.
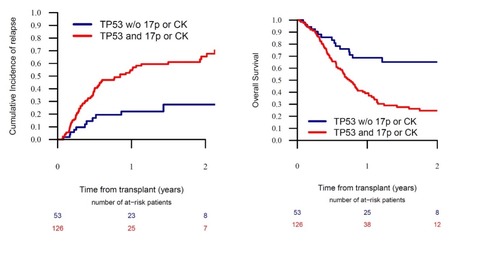
In pts with mutated TP53 AML, differences in conditioning intensity, donor source and GVHD prophylaxis strategy did not significantly affect OS or CIR. However, in pts with TP53 mutant AML the presence of either 17p abnormalities (abn17p) or complex karyotype (CK) provided further prognostic value. In pts with mutated TP53 AML without abn17p or CK, the 2-year OS was 65.2% [95%CI: 48.4-77.6]; in comparison, in pts with mutated TP53 AML with either abn17p or CK, the OS was significantly lower (24.6% [95%CI: 16.2-34], p=0.001) (figure). In a multivariate analysis the additive poor prognostic effect of abn17p and CK on the OS of pts with mutated TP53 AML was maintained. In pts with mutant TP53 AML with either abn17p or CK, the leukaemia free survival (LFS) at 2 years was 15.2% ([95% CI: 8-24.6], p=0.001), which was lower than that in pts with mutated TP53 AML, but without abn17p or CK, (LFS: 59.9% [95%CI: 41.7-74.1]). In pts with mutant TP53 AML with either abn17p or CK, the CIR was 65.4% ([95% CI: 53.9-74.8], p=0.001), which was higher than that in pts with mutated TP53 AML, but without abn17p or CK, (CIR: 27.5% [95% CI: 13.4-43.7]) (figure).
Conclusion
The adverse prognostic effect of TP53 mutations in AML is only evident in pts with co-occurring abn17p or CK. Even in pts with mutated TP53 AML and a co-occurring abn17p or CK, a significant proportion of pts achieved long-term survival. Our data is practice changing and assists in the identification of pts who stand to benefit from an allo-SCT. Finally, the enrichment of abn17p in pts with TP53 mutations, and their additional poor prognostic implication, supports the biological importance of biallelic loss of TP53 activity in AML.
Keyword(s): Acute myeloid leukemia, Genomics, Stem cell transplant
Abstract: S232
Type: Oral Presentation
Session title: Stem cell transplantation - Preclinical/experimental
Background
TP53 mutations in patients (pts) with acute myeloid leukaemia (AML) has been associated with an unfavourable outcome including after allogeneic stem cell transplantation (allo-SCT). This has led some to question the benefit of allo-SCT for TP53 mutant AML, although it has not been studied in a large patient cohort.
Aims
To identify factors which influence transplant outcome in pts with mutant TP53 AML.
Methods
We identified pts with AML in first complete remission, and with either intermediate or adverse risk cytogenetics, whose TP53 mutation status was reported to the EBMT registry. All ages, conditioning intensity or donor source were included.
Results
We identified 179 pts with AML with TP53 mutations and compared their characteristics and outcomes to 601 pts with AML without TP53 mutations. Pts were transplanted between 2015-2019. Chromosomal losses in chromosomes 17p (31% vs. 3%), 5q (49% vs. 5%), and 7q (34% vs. 8%) as well as complex (65% vs. 12%) and monosomal karyotypes (41% vs. 8%) were significantly increased in the cohort of pts with mutated TP53, in contrast to those with unmutated TP53.
The overall survival (OS) at 2 years was significantly impaired in pts with mutated TP53 as compared to the cohort without (mutated TP53, 35.1% [95% CI: 26.7-43.7]; without TP53 mutations, 64% [95% CI: 59.1-68.4], p= 0.001). This was a result of an increased cumulative incidence of relapse (CIR) at 2 years in pts with mutated TP53 as compared to the cohort without mutated TP53 (mutated TP53, 55% [95% CI: 45.2-63.8]; without TP53 mutations, 25.2% [95%CI: 21.2-29.3], p= 0.001). The adverse prognostic impact of TP53 mutations on OS (HR=1.5, 95%CI 1.1-2.1) and CIR (HR=1.4, 95%CI: 1.2-1.9) was maintained in a multivariate model that accounted for the adverse prognostic value of other known cytogenetic, and transplant, variables.
In pts with mutated TP53 AML, differences in conditioning intensity, donor source and GVHD prophylaxis strategy did not significantly affect OS or CIR. However, in pts with TP53 mutant AML the presence of either 17p abnormalities (abn17p) or complex karyotype (CK) provided further prognostic value. In pts with mutated TP53 AML without abn17p or CK, the 2-year OS was 65.2% [95%CI: 48.4-77.6]; in comparison, in pts with mutated TP53 AML with either abn17p or CK, the OS was significantly lower (24.6% [95%CI: 16.2-34], p=0.001) (figure). In a multivariate analysis the additive poor prognostic effect of abn17p and CK on the OS of pts with mutated TP53 AML was maintained. In pts with mutant TP53 AML with either abn17p or CK, the leukaemia free survival (LFS) at 2 years was 15.2% ([95% CI: 8-24.6], p=0.001), which was lower than that in pts with mutated TP53 AML, but without abn17p or CK, (LFS: 59.9% [95%CI: 41.7-74.1]). In pts with mutant TP53 AML with either abn17p or CK, the CIR was 65.4% ([95% CI: 53.9-74.8], p=0.001), which was higher than that in pts with mutated TP53 AML, but without abn17p or CK, (CIR: 27.5% [95% CI: 13.4-43.7]) (figure).

Conclusion
The adverse prognostic effect of TP53 mutations in AML is only evident in pts with co-occurring abn17p or CK. Even in pts with mutated TP53 AML and a co-occurring abn17p or CK, a significant proportion of pts achieved long-term survival. Our data is practice changing and assists in the identification of pts who stand to benefit from an allo-SCT. Finally, the enrichment of abn17p in pts with TP53 mutations, and their additional poor prognostic implication, supports the biological importance of biallelic loss of TP53 activity in AML.
Keyword(s): Acute myeloid leukemia, Genomics, Stem cell transplant


