
Contributions
Abstract: S162
Type: Oral Presentation
Session title: Correlating mutations with phenotype in MDS
Background
Myelodysplastic syndromes (MDS) are a group of heterogeneous clonal disorders essentially occurring in elderly individuals and associated with a high risk of progression to acute myeloid leukemia (AML). A majority of MDS patients harbors somatic mutations including in genes encoding for epigenetic regulators such as TET2 and IDH1 and 2. Those mutations have been also found in individuals with clonal hematopoiesis of indeterminate potential (CHIP), a preleukemic state. However, mechanisms of disease progression remain largely unknown and may implicate defects in the anti-leukemic immunosurveillance. Among immune cells, Natural Killer (NK) cells are key players in charge of eliminating tumor cells. At diagnosis of AML and MDS, we and others showed impaired NK cell's phenotype and function with a reduction of cytotoxicity and cytokine release (Khaznadar et al. 2015 J Immunol, Carlsten et al. 2010 Leukemia). However no data are yet available on the impact on NK-cells of the mutation status in MDS patients at diagnosis.
Aims
We hypothesized that MDS patients with TET2/IDH mutations could show particular defective NK cell profile leading to the disruption of their anti-leukemic activity, by contrast to other MDS-related mutations.
Methods
Bone marrow (BM) and blood samples from 63 patients with MDS were collected, from the Hematological Departments of Saint Louis Hospital at diagnosis prior any treatment. Next generation sequencing (NGS) using a 80 genes panel was performed on sorted blood NK and T cells. NK cells phenotype was analyzed by flow-cytometry and NK-cell mediated cytotoxicity against the leukemic K562 cell-line was evaluated in calcein-release assays. The NK-cell methylation landscape has been assessed by representation bisulfite sequencing (RRBS). Finally, NK-cells treated in vitro with HMA have been analyzed for NK receptor expression before and after HMA exposure.
Results
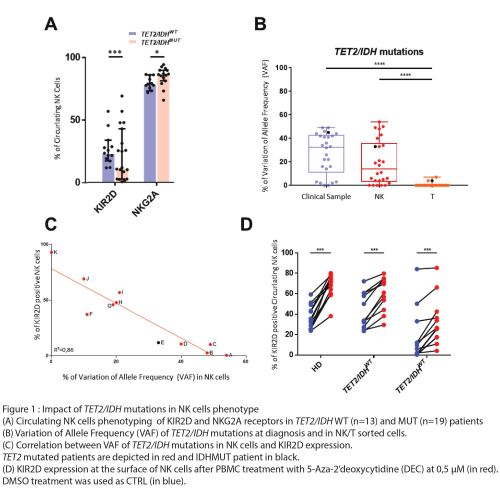
Phenotyping of NK cells in blood and BM of MDS patients showed a strong reduction of KIR2D receptors expression counterbalanced by the increase of NKG2A at the NK-cell surface in patients harboring TET2 or IDH1/2 mutations (Figure 1A). The NGS analysis revealed a shared mutation profile between blasts and NK cells, but not T lymphocytes, in the MDS patients (Figure 1B). Interestingly, KIR expression correlated with the Variation of Allele Frequency (VAF) of TET2 mutations in NK cells (Figure 1C). Chromatin immuno-precipitation experiments in primary NK cells unveiled the direct binding of TET2 on the KIR promoter region. In parallel, RRBS analysis showed DNA hypermethylation of the KIR locus and other genes involved in NK cell physiology, including PRF1 and IFNG, in the TET2MUT NK-cells by comparison to TET2WT NK-cells. Importantly, in vitro HMA treatment of NK-cells restores KIR2D cell-surface expression (figure 1D).
Conclusion
Altogether, our findings showed that mutations in TET2 altered NK-cell biology in MDS patients. This study highlights the potential dual role of mutations in the emergence of MDS and immune defects, which could lead to progression to AML by associating oncogenic mechanisms and tumor escape to immunosurveillance.
Keyword(s): Methylation, Myelodysplasia, NK cell, Tumor immunology
Abstract: S162
Type: Oral Presentation
Session title: Correlating mutations with phenotype in MDS
Background
Myelodysplastic syndromes (MDS) are a group of heterogeneous clonal disorders essentially occurring in elderly individuals and associated with a high risk of progression to acute myeloid leukemia (AML). A majority of MDS patients harbors somatic mutations including in genes encoding for epigenetic regulators such as TET2 and IDH1 and 2. Those mutations have been also found in individuals with clonal hematopoiesis of indeterminate potential (CHIP), a preleukemic state. However, mechanisms of disease progression remain largely unknown and may implicate defects in the anti-leukemic immunosurveillance. Among immune cells, Natural Killer (NK) cells are key players in charge of eliminating tumor cells. At diagnosis of AML and MDS, we and others showed impaired NK cell's phenotype and function with a reduction of cytotoxicity and cytokine release (Khaznadar et al. 2015 J Immunol, Carlsten et al. 2010 Leukemia). However no data are yet available on the impact on NK-cells of the mutation status in MDS patients at diagnosis.
Aims
We hypothesized that MDS patients with TET2/IDH mutations could show particular defective NK cell profile leading to the disruption of their anti-leukemic activity, by contrast to other MDS-related mutations.
Methods
Bone marrow (BM) and blood samples from 63 patients with MDS were collected, from the Hematological Departments of Saint Louis Hospital at diagnosis prior any treatment. Next generation sequencing (NGS) using a 80 genes panel was performed on sorted blood NK and T cells. NK cells phenotype was analyzed by flow-cytometry and NK-cell mediated cytotoxicity against the leukemic K562 cell-line was evaluated in calcein-release assays. The NK-cell methylation landscape has been assessed by representation bisulfite sequencing (RRBS). Finally, NK-cells treated in vitro with HMA have been analyzed for NK receptor expression before and after HMA exposure.
Results
Phenotyping of NK cells in blood and BM of MDS patients showed a strong reduction of KIR2D receptors expression counterbalanced by the increase of NKG2A at the NK-cell surface in patients harboring TET2 or IDH1/2 mutations (Figure 1A). The NGS analysis revealed a shared mutation profile between blasts and NK cells, but not T lymphocytes, in the MDS patients (Figure 1B). Interestingly, KIR expression correlated with the Variation of Allele Frequency (VAF) of TET2 mutations in NK cells (Figure 1C). Chromatin immuno-precipitation experiments in primary NK cells unveiled the direct binding of TET2 on the KIR promoter region. In parallel, RRBS analysis showed DNA hypermethylation of the KIR locus and other genes involved in NK cell physiology, including PRF1 and IFNG, in the TET2MUT NK-cells by comparison to TET2WT NK-cells. Importantly, in vitro HMA treatment of NK-cells restores KIR2D cell-surface expression (figure 1D).

Conclusion
Altogether, our findings showed that mutations in TET2 altered NK-cell biology in MDS patients. This study highlights the potential dual role of mutations in the emergence of MDS and immune defects, which could lead to progression to AML by associating oncogenic mechanisms and tumor escape to immunosurveillance.
Keyword(s): Methylation, Myelodysplasia, NK cell, Tumor immunology


