
Contributions
Abstract: PB1802
Type: Publication Only
Session title: Stem cell transplantation - Experimental
Background
Allogeneic hematopoietic cell transplantation (allo-HSCT) is the most effective curative therapy in patients with acute myeloid leukemia (AML). However, relapse from the original disease is still one of the main causes of treatment failure. Thus high-sensitivity methods to detect minimal residual disease (MRD) in AML after transplantation are crucial. Single cell sequencing indicated that higher clonal diversity at the stem/progenitor cell level (HSPC) in AML, but the significance of MRD monitoring at the HSPC level is unclear.
Aims
To evaluate the value of MRD monitoring based on the stem cell level in predicting post-transplantation relapse.
Methods
DNA was extracted from cells collected from bone marrow (BM) samples. Genetic profiling included the targeted sequencing of 37 genes, which had been selected based on their known or potential involvement in the pathogenesis of acute myelogenous leukemia. Multiplexed libraries were sequenced with Illumina NovaSeq6000 and average raw sequencing depth on target per sample ≥10000×. The mutation variant allele frequency (VAF) ≥1% was considered as MRD positive criterion.
Results
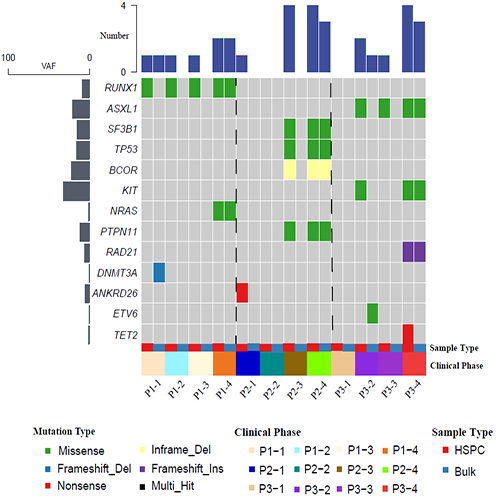
We performed next-generation sequencing (NGS) to profile the mutation status in three AML patients (P1-P3) with relapse after transplantation. For each patient, two BM samples including bulk BM and CD34+ HSPC were collected at post-HSCT at day30、day60、day90 and relapse, respectively. Compared to bulk BM and multidimensional flow cytometry (MFC), MRD monitoring based on the HSPC level is likely more sensitive to predict relapse after transplantation (Figure 1). One of the patients was detectable as MRD positive at post-HSCT day30. Moreover, we observed that some hotspot mutations might be detected only in the HSPC sample.
Conclusion
Our study unveiled that phenotypically normal CD34+ HSPC possess some genetic abnormalities in cases with negative MCF MRD. NGS-defined MRD at the HSPC level in AML patients is feasible and can predict post-transplantation relapse much earlier.
Keyword(s): AML, MRD, Relapse
Abstract: PB1802
Type: Publication Only
Session title: Stem cell transplantation - Experimental
Background
Allogeneic hematopoietic cell transplantation (allo-HSCT) is the most effective curative therapy in patients with acute myeloid leukemia (AML). However, relapse from the original disease is still one of the main causes of treatment failure. Thus high-sensitivity methods to detect minimal residual disease (MRD) in AML after transplantation are crucial. Single cell sequencing indicated that higher clonal diversity at the stem/progenitor cell level (HSPC) in AML, but the significance of MRD monitoring at the HSPC level is unclear.
Aims
To evaluate the value of MRD monitoring based on the stem cell level in predicting post-transplantation relapse.
Methods
DNA was extracted from cells collected from bone marrow (BM) samples. Genetic profiling included the targeted sequencing of 37 genes, which had been selected based on their known or potential involvement in the pathogenesis of acute myelogenous leukemia. Multiplexed libraries were sequenced with Illumina NovaSeq6000 and average raw sequencing depth on target per sample ≥10000×. The mutation variant allele frequency (VAF) ≥1% was considered as MRD positive criterion.
Results
We performed next-generation sequencing (NGS) to profile the mutation status in three AML patients (P1-P3) with relapse after transplantation. For each patient, two BM samples including bulk BM and CD34+ HSPC were collected at post-HSCT at day30、day60、day90 and relapse, respectively. Compared to bulk BM and multidimensional flow cytometry (MFC), MRD monitoring based on the HSPC level is likely more sensitive to predict relapse after transplantation (Figure 1). One of the patients was detectable as MRD positive at post-HSCT day30. Moreover, we observed that some hotspot mutations might be detected only in the HSPC sample.

Conclusion
Our study unveiled that phenotypically normal CD34+ HSPC possess some genetic abnormalities in cases with negative MCF MRD. NGS-defined MRD at the HSPC level in AML patients is feasible and can predict post-transplantation relapse much earlier.
Keyword(s): AML, MRD, Relapse


