
Contributions
Abstract: PB1490
Type: Publication Only
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Tumor B-cells in chronic lymphocytic leukemia possess extreme narrowing of the IGHV gene repertoire comparing to other lymphoproliferative diseases. B-cell receptors with a highly homologous structure (SARs) - account for more than 40% of all CLL cases, with 29 such most common subgroups accounting for more than 13% of all CLL cases. CLL#1, CLL#2, and CLL#8 SARs are associated with an extremely aggressive course of the disease, while CLL#4 - with indolent. It was also suggested that tumor cells expressing certain SARs may also differ in the frequency and patterns of genetic lesions.
Aims
To determine the frequency of TP53, NOTCH1, and SF3B1 gene mutations in CLL#1, CLL#6, CLL#3, CLL#99, and CLL#28A subgroups of patients from the Russian cohort.
Methods
The study included 88 CLL patients admitted to the National Research Center for Hematology (Moscow, Russia) from 2012 to 2020. Rearranged IGHV gene sequences and SARs were determined according to the procedure recommended by ERIC. NOTCH1 gene (exon 34) mutations were studied by fragment analysis or by NGS as described by Campregher et al. TP53 gene mutations (exons 2-11 exons) were determined by NGS according to Pavlova et al. SF3B1 gene (exons 14-16) mutations were assessed by NGS.
Results
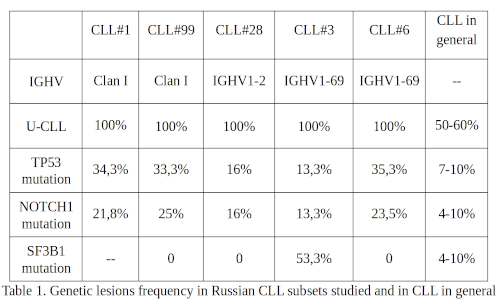
Peripheral blood DNA samples from CLL#1 (N=32), CLL#6 (N=17), CLL#3 (N=15), CLL#99 (N=12), and CLL#28A (N=12) patients were studied. TP53 gene mutations were detected with a high frequency in the CLL #6, CLL#1, CLL#99, and CLL#28A subgroups in 6 (35.3%), 11 (34.3%), 4 (33.3%) and 2 (16%) cases respectively. It should be noted that in all patients with CLL#99 and CLL#28A, only minor VAF (<5%) mutations were found. Most CLL#1 and CLL#6 patients have VAF>10%. Two patients with CLL#6 and one with CLL#99 had 2 clones simultaneously. In the CLL#3 subgroup, only 2 cases with mutations in this gene were identified, however one had 2 clones, and the other - 4. Mutations in the NOTCH1 gene were detected in patients with CLL#99, CLL#6, CLL#1, CLL#28A, and CLL#3 with a frequency of 3 (25%), 4 (23.5%), 7 (21.8%), 2 (16%) and 2 (13%) respectively. In most cases, the widespread c.7544_7545delCT mutation was detected. Also, c.7558_7561delCTTC and p.Q2444X mutations were found in two cases with CLL#1 and c.7547_7551del deletion in a patient with CLL#28A. SF3B1 gene mutations were detected in 8 (53.3%) CLL#3 patients. In CLL#6 (11 patients studied), CLL#99, and CLL#28A no SF3B1 gene mutations were found and in CLL#1 subgroup these mutations have not been assessed yet.
Conclusion
Here we present the data concerning the higher frequency of mutations in CLL#1, CLL#6, and CLL#3 subgroups than have been reported so far. Perhaps this is since some of our patients were tested after they already received FCR treatment and subsequently relapsed. It should be noted that CDR3 in subgroups CLL#1 and #99 have a similar motif, differ only by 1 amino acid, and are formed using the IGHV genes of the clan 1 (mostly IGHV1-2). CLL#28A is also based on IGHV1-2, and CLL#3 and CLL#6 are based on IGHV1-69. In the CLL#99 subgroup, as well as in the CLL#1 subgroup of a similar receptor structure, mutations in the TP53 gene are abnormally common, but with low VAF, which currently has no proven negative clinical significance. Interestingly, the two SARS subgroups with the IGHV1-69 gene usage demonstrate an opposite TP53, NOTCH1, and SF3B1 gene mutation pattern. Further study of genetic lesions in diverse SARs subgroups CLL patients should help to improve understanding of the pathogenesis, prognostication, and therapy of the disease.
Keyword(s): IgH rearrangment, Mutation analysis, Notch, P53
Abstract: PB1490
Type: Publication Only
Session title: Chronic lymphocytic leukemia and related disorders - Biology & Translational Research
Background
Tumor B-cells in chronic lymphocytic leukemia possess extreme narrowing of the IGHV gene repertoire comparing to other lymphoproliferative diseases. B-cell receptors with a highly homologous structure (SARs) - account for more than 40% of all CLL cases, with 29 such most common subgroups accounting for more than 13% of all CLL cases. CLL#1, CLL#2, and CLL#8 SARs are associated with an extremely aggressive course of the disease, while CLL#4 - with indolent. It was also suggested that tumor cells expressing certain SARs may also differ in the frequency and patterns of genetic lesions.
Aims
To determine the frequency of TP53, NOTCH1, and SF3B1 gene mutations in CLL#1, CLL#6, CLL#3, CLL#99, and CLL#28A subgroups of patients from the Russian cohort.
Methods
The study included 88 CLL patients admitted to the National Research Center for Hematology (Moscow, Russia) from 2012 to 2020. Rearranged IGHV gene sequences and SARs were determined according to the procedure recommended by ERIC. NOTCH1 gene (exon 34) mutations were studied by fragment analysis or by NGS as described by Campregher et al. TP53 gene mutations (exons 2-11 exons) were determined by NGS according to Pavlova et al. SF3B1 gene (exons 14-16) mutations were assessed by NGS.
Results
Peripheral blood DNA samples from CLL#1 (N=32), CLL#6 (N=17), CLL#3 (N=15), CLL#99 (N=12), and CLL#28A (N=12) patients were studied. TP53 gene mutations were detected with a high frequency in the CLL #6, CLL#1, CLL#99, and CLL#28A subgroups in 6 (35.3%), 11 (34.3%), 4 (33.3%) and 2 (16%) cases respectively. It should be noted that in all patients with CLL#99 and CLL#28A, only minor VAF (<5%) mutations were found. Most CLL#1 and CLL#6 patients have VAF>10%. Two patients with CLL#6 and one with CLL#99 had 2 clones simultaneously. In the CLL#3 subgroup, only 2 cases with mutations in this gene were identified, however one had 2 clones, and the other - 4. Mutations in the NOTCH1 gene were detected in patients with CLL#99, CLL#6, CLL#1, CLL#28A, and CLL#3 with a frequency of 3 (25%), 4 (23.5%), 7 (21.8%), 2 (16%) and 2 (13%) respectively. In most cases, the widespread c.7544_7545delCT mutation was detected. Also, c.7558_7561delCTTC and p.Q2444X mutations were found in two cases with CLL#1 and c.7547_7551del deletion in a patient with CLL#28A. SF3B1 gene mutations were detected in 8 (53.3%) CLL#3 patients. In CLL#6 (11 patients studied), CLL#99, and CLL#28A no SF3B1 gene mutations were found and in CLL#1 subgroup these mutations have not been assessed yet.

Conclusion
Here we present the data concerning the higher frequency of mutations in CLL#1, CLL#6, and CLL#3 subgroups than have been reported so far. Perhaps this is since some of our patients were tested after they already received FCR treatment and subsequently relapsed. It should be noted that CDR3 in subgroups CLL#1 and #99 have a similar motif, differ only by 1 amino acid, and are formed using the IGHV genes of the clan 1 (mostly IGHV1-2). CLL#28A is also based on IGHV1-2, and CLL#3 and CLL#6 are based on IGHV1-69. In the CLL#99 subgroup, as well as in the CLL#1 subgroup of a similar receptor structure, mutations in the TP53 gene are abnormally common, but with low VAF, which currently has no proven negative clinical significance. Interestingly, the two SARS subgroups with the IGHV1-69 gene usage demonstrate an opposite TP53, NOTCH1, and SF3B1 gene mutation pattern. Further study of genetic lesions in diverse SARs subgroups CLL patients should help to improve understanding of the pathogenesis, prognostication, and therapy of the disease.
Keyword(s): IgH rearrangment, Mutation analysis, Notch, P53


