
Contributions
Abstract: PB1356
Type: Publication Only
Session title: Acute lymphoblastic leukemia - Biology & Translational Research
Background
BCR-ABL is a constitutively active tyrosine kinase formed by the t(9;22) reciprocal translocation, with the p190 transcript being the most frequent variant in Ph+ ALL¹. Tyrosine kinase inhibitor (TKI) resistance develops in a significant number of Ph+ ALL patients over the course of treatment²,³,⁴. As 15-20% of children, and about half of adults treated for Ph+ ALL ultimately relapse, there is need for a more sensitive and easier to use molecular test with a shorter time-to-result (TTR) for the monitoring of MRD and treatment response in p190-positive patients³,⁵,⁶.
Aims
The objective of this study was to compare the performance of the novel Cepheid Xpert BCR-ABL Ultra p190 Prototype Assay to the Asuragen QuantideX® qPCR BCR-ABL minor Kit, the Qiagen ipsogen® BCR-ABL1 mbcr Kit, and an in-house developed reverse transcriptase digital droplet PCR (RT-ddPCR) assay using samples contrived using Philadelphia chromosome-positive (Ph+) ALL patient peripheral blood. Our goal was to create a fast and easy to use quantitative assay with higher sensitivity than currently available assays for use in the monitoring of treatment efficacy and minimal residual disease (MRD) in Ph+ ALL patients.
Methods
Three p190-positive ALL total RNA samples were spiked into p190-negative EDTA whole blood lysate matrix diluents and serially diluted to target %BCR-ABL p190/ABL outputs from approximately 25% (LR [log reduction] 0.60) to near the assays expected limit of detection (LoD), approximately 0.005% (LR 4.30). A subset of each level underwent sample preparation, total RNA extraction, and RT-qPCR all in the Cepheid GeneXpert automated cartridge system, and another subset had total RNA extracted using the Zymo Direct-zol™ RNA Miniprep Plus Kit. RNA was qualified and quantified using the Thermo Scientific NanoDrop™ 2000 Spectrophotometer before being tested per the comparator assay vendors’ recommendations. The Cepheid Prototype assay utilized 4.5mL of lysate, whereas the Asuragen assay used 1-2 µg of RNA per test, and the Qiagen and RT-ddPCR assays used 1 µg of RNA per test.
The Cepheid Prototype assay was evaluated against the three comparator assays using Deming regressions with regression confidence intervals determined using jackknife resampling. Bland-Altman plots were utilized to evaluate bias between assay quantitation outputs. Hit-rates were assessed to compare the LoD between assays.
Results
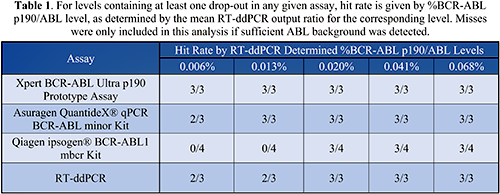
Deming regressions and associated Bland-Altman plots showed a high degree of correlation and agreement between comparator assays and the Cepheid Prototype assay. Replicates that did not detect p190 were not included in this analysis. Dropout rates at low target %BCR-ABL p190/ABL levels were compared between assays. The Cepheid Prototype assay had the best performance in terms of hit-rate, followed by the Asuragen, RT-ddPCR, and Qiagen assays (Table 1).
Conclusion
The novel Cepheid Prototype assay showed a high level of agreement with the three comparator assays and gave the lowest LoD overall in this study using samples contrived from Ph+ ALL patient peripheral blood. TTR was minimized in the Cepheid Prototype assay to 130 minutes, while the comparator assays all had a TTR of approximately five hours or greater. While RNA extraction is performed within Cepheid Prototype assay cartridge and is included in the 130-minute TTR, the comparator assays require additional expensive and labor-intensive extraction steps that can last one-hour to multiple days, which are not included in the five-hour TTR estimate.
Keyword(s): Acute lymphoblastic leukemia, Assay, Philadelphia chromosome, RT-PCR
Abstract: PB1356
Type: Publication Only
Session title: Acute lymphoblastic leukemia - Biology & Translational Research
Background
BCR-ABL is a constitutively active tyrosine kinase formed by the t(9;22) reciprocal translocation, with the p190 transcript being the most frequent variant in Ph+ ALL¹. Tyrosine kinase inhibitor (TKI) resistance develops in a significant number of Ph+ ALL patients over the course of treatment²,³,⁴. As 15-20% of children, and about half of adults treated for Ph+ ALL ultimately relapse, there is need for a more sensitive and easier to use molecular test with a shorter time-to-result (TTR) for the monitoring of MRD and treatment response in p190-positive patients³,⁵,⁶.
Aims
The objective of this study was to compare the performance of the novel Cepheid Xpert BCR-ABL Ultra p190 Prototype Assay to the Asuragen QuantideX® qPCR BCR-ABL minor Kit, the Qiagen ipsogen® BCR-ABL1 mbcr Kit, and an in-house developed reverse transcriptase digital droplet PCR (RT-ddPCR) assay using samples contrived using Philadelphia chromosome-positive (Ph+) ALL patient peripheral blood. Our goal was to create a fast and easy to use quantitative assay with higher sensitivity than currently available assays for use in the monitoring of treatment efficacy and minimal residual disease (MRD) in Ph+ ALL patients.
Methods
Three p190-positive ALL total RNA samples were spiked into p190-negative EDTA whole blood lysate matrix diluents and serially diluted to target %BCR-ABL p190/ABL outputs from approximately 25% (LR [log reduction] 0.60) to near the assays expected limit of detection (LoD), approximately 0.005% (LR 4.30). A subset of each level underwent sample preparation, total RNA extraction, and RT-qPCR all in the Cepheid GeneXpert automated cartridge system, and another subset had total RNA extracted using the Zymo Direct-zol™ RNA Miniprep Plus Kit. RNA was qualified and quantified using the Thermo Scientific NanoDrop™ 2000 Spectrophotometer before being tested per the comparator assay vendors’ recommendations. The Cepheid Prototype assay utilized 4.5mL of lysate, whereas the Asuragen assay used 1-2 µg of RNA per test, and the Qiagen and RT-ddPCR assays used 1 µg of RNA per test.
The Cepheid Prototype assay was evaluated against the three comparator assays using Deming regressions with regression confidence intervals determined using jackknife resampling. Bland-Altman plots were utilized to evaluate bias between assay quantitation outputs. Hit-rates were assessed to compare the LoD between assays.
Results
Deming regressions and associated Bland-Altman plots showed a high degree of correlation and agreement between comparator assays and the Cepheid Prototype assay. Replicates that did not detect p190 were not included in this analysis. Dropout rates at low target %BCR-ABL p190/ABL levels were compared between assays. The Cepheid Prototype assay had the best performance in terms of hit-rate, followed by the Asuragen, RT-ddPCR, and Qiagen assays (Table 1).

Conclusion
The novel Cepheid Prototype assay showed a high level of agreement with the three comparator assays and gave the lowest LoD overall in this study using samples contrived from Ph+ ALL patient peripheral blood. TTR was minimized in the Cepheid Prototype assay to 130 minutes, while the comparator assays all had a TTR of approximately five hours or greater. While RNA extraction is performed within Cepheid Prototype assay cartridge and is included in the 130-minute TTR, the comparator assays require additional expensive and labor-intensive extraction steps that can last one-hour to multiple days, which are not included in the five-hour TTR estimate.
Keyword(s): Acute lymphoblastic leukemia, Assay, Philadelphia chromosome, RT-PCR


