
Contributions
Abstract: PB2434
Type: Publication Only
Background
Chimerism analysis plays a key role in the detection of bone marrow engraftment after allogeneic bone marrow transplantation (BMT). The analysis of short tandem repeats (STR) loci from the donor and the recipient DNA and calculation of individual STR markers percentage is one of routine test for chimerism monitoring. One of most often complications are the stutter PCR peaks appearance due to polymerase slippage during amplification of target STRs. The stutter peak may concur with an informative recipient`s marker hindering chimerism estimation based on that locus. This problem seems to be especially serious in case of a sex-matched sibling bone marrow transplantation. Thereby, the absence of “stutter-peaks free” markers hinders mixed chimerism estimation at the point of low recipient hematopoiesis output. Previously we demonstrated the possibility of the universal formulas correction for the chimerism calculation excluding stutter percentage.
Aims
Detection of low levels of mixed chimerism is possible if only recipient peak is greater then expected donor stutter peak plus some threshold because of error of measurement. The aim of this work was to evaluate the portions of variation of peak level caused by person (donor), locus and noise of measurements and then to calculate the threshold according given type II error of stutter-complicated chimerism detection.
Methods
Data includes measurements of stutter peaks (SP) percentage in 10 loci in DNA samples of 23 donor/recipient sets . In each donor/patient set first measurement was done for donor then further stutter percentage was calculated on the DNA samples of patients with verified complete donor chimerism in 6 up to 14 time points. Additionally we made measurements in parallel probes – 8 aliquots for 8 donors in 7 loci in the most recent time point samples. The UNIVARIATE and GLM SAS procedures were used to calculate simple statistics and run analysis of variance.
Results
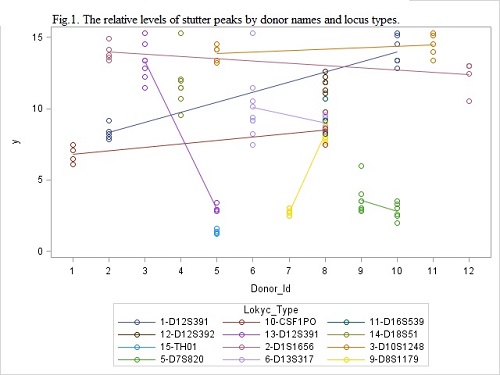
Relative levels of stutter peaks by locus types for 23 donor/patient sets is shown on Fig.1, multiple points means repeated measurements. The analysis of variance shows that the main sources of variation are the person (F=5.4, p<.0001) and locus(F=12.8, p<.0001) but not time (number) of measurement (F=0.77, p=0.6). We also compared the variance of repeated measurement of SP of donor/patient set and variance in aliquots of last patient’s measurements . At that case the analysis of variance shows that the main sources of variation are the person (F=49.45, p<.0001) and locus type (F=45.16, p<.0001) but not aliquots (number of the parallel probes) (F=1.93, p=0.08). The deviations in repeated measurement and in aliquots are close (0.73 vs 0.54) and we can regard them as measurement errors. Then we estimate the distribution of errors, for this we take regression residuals (RR) after fitting by donors and locus types. The mean of RR is zero by definition, the standard deviation is 0.73, 95% percentile is 0.86. The threshold for chimerism detection with 5% of type II error can be calculated by formula: Th5=SP+0.86, where SP is value of stutter peak percentage of donor.
Conclusion
We recognized that the stutter peaks are variated mostly by locus type and by person but rather stable in series of measurements in different time points and in several aliquots for the same locus. That means that we can construct the individual chimerism detection rule using donor SP and measurement error which we have evaluated.
Session topic: 23. Stem cell transplantation - Clinical
Keyword(s): Donor, STR-PCR, Transplant, Chimerism
Abstract: PB2434
Type: Publication Only
Background
Chimerism analysis plays a key role in the detection of bone marrow engraftment after allogeneic bone marrow transplantation (BMT). The analysis of short tandem repeats (STR) loci from the donor and the recipient DNA and calculation of individual STR markers percentage is one of routine test for chimerism monitoring. One of most often complications are the stutter PCR peaks appearance due to polymerase slippage during amplification of target STRs. The stutter peak may concur with an informative recipient`s marker hindering chimerism estimation based on that locus. This problem seems to be especially serious in case of a sex-matched sibling bone marrow transplantation. Thereby, the absence of “stutter-peaks free” markers hinders mixed chimerism estimation at the point of low recipient hematopoiesis output. Previously we demonstrated the possibility of the universal formulas correction for the chimerism calculation excluding stutter percentage.
Aims
Detection of low levels of mixed chimerism is possible if only recipient peak is greater then expected donor stutter peak plus some threshold because of error of measurement. The aim of this work was to evaluate the portions of variation of peak level caused by person (donor), locus and noise of measurements and then to calculate the threshold according given type II error of stutter-complicated chimerism detection.
Methods
Data includes measurements of stutter peaks (SP) percentage in 10 loci in DNA samples of 23 donor/recipient sets . In each donor/patient set first measurement was done for donor then further stutter percentage was calculated on the DNA samples of patients with verified complete donor chimerism in 6 up to 14 time points. Additionally we made measurements in parallel probes – 8 aliquots for 8 donors in 7 loci in the most recent time point samples. The UNIVARIATE and GLM SAS procedures were used to calculate simple statistics and run analysis of variance.
Results
Relative levels of stutter peaks by locus types for 23 donor/patient sets is shown on Fig.1, multiple points means repeated measurements. The analysis of variance shows that the main sources of variation are the person (F=5.4, p<.0001) and locus(F=12.8, p<.0001) but not time (number) of measurement (F=0.77, p=0.6). We also compared the variance of repeated measurement of SP of donor/patient set and variance in aliquots of last patient’s measurements . At that case the analysis of variance shows that the main sources of variation are the person (F=49.45, p<.0001) and locus type (F=45.16, p<.0001) but not aliquots (number of the parallel probes) (F=1.93, p=0.08). The deviations in repeated measurement and in aliquots are close (0.73 vs 0.54) and we can regard them as measurement errors. Then we estimate the distribution of errors, for this we take regression residuals (RR) after fitting by donors and locus types. The mean of RR is zero by definition, the standard deviation is 0.73, 95% percentile is 0.86. The threshold for chimerism detection with 5% of type II error can be calculated by formula: Th5=SP+0.86, where SP is value of stutter peak percentage of donor.

Conclusion
We recognized that the stutter peaks are variated mostly by locus type and by person but rather stable in series of measurements in different time points and in several aliquots for the same locus. That means that we can construct the individual chimerism detection rule using donor SP and measurement error which we have evaluated.
Session topic: 23. Stem cell transplantation - Clinical
Keyword(s): Donor, STR-PCR, Transplant, Chimerism


