
Contributions
Abstract: PB1894
Type: Publication Only
Background
One of the most controversial causes of TKI resistance in patients with CML is the pathogenic BCR- ABL del. c.1086-1270 transcript. This transcript was first described in 2008 in TKI resistant patients (Curvo et al., 2008). BCR-ABL del. c.1086-1270 transcript encodes truncated fusion protein BCR-ABL p.R362fs*21 which is an alternative splicing product. It has been shown that such truncated protein is unlikely to have tyrosine kinase activity because of the disfunction of the ATP-binding site (Meggyesi et al., 2012). However, we suggest that pathogenic effect of BCR-ABL p.R362fs*21 expression is possible due to the dimerization with a typical protein BCR-ABL p210. Such mechanism has already been proved for the splice variant of serine-threonine kinase BRAF (Poulikakos etal., 2011).
Aims
To evaluate the pathogenic effect of BCR-ABL del. c.1086-1270 (p.R362fs*21) on TKI resistance formation in Russian patients with CML.
Methods
33 male and 50 female CML patients (age 24-80) with BCR-ABL transcript level > 0.1% were included in the study. BCR-ABL exon 7 deletion was analyzed with two round (nested) PCR followed by Sanger sequencing. Initial screening for deletions was performed by means of fragment analysis (Applied Biosystems 3130).
Results
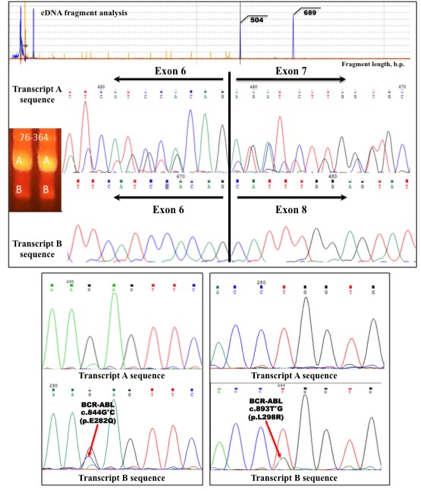
Fragment analysis detected BCR-ABL del. c.1086-1270 transcript in 32 patients (38%). The ratio of TKI-sensitive to TKI-resistant among these 32 patients was approximately 47% (15 patients) to 53% (17 patients) respectively. Direct sequencing showed the tumor clone, which contains the BCR-ABL1 del.c.1086-1270 always paired with a normal clone of BCR-ABL p210, in all TKI-resistant patients. This fact was verified by the results of agarose gel electrophoresis. We suggest, that these results show an indirect evidence of the dimerization of the protein BCR-ABL del. c.1086-1270 with chimeric protein Bcr-Abl p210. Furthermore, the deletion clone of each TKI-resistant patient always has a point mutation (F317V, F317L, E282Q, M351T, T315I) which is unlikely for the normal clone (Figure 1). This fact contradicts the alternative splicing theory.
Conclusion
Our results show a definite correlation between the presence of the deletion and TKI-resistance. BCR- ABL1 del.c.1086-1270 is likely to play a trigger-role in TKI-resistance formation: clones with the exon 7 deletion are extremely prone to the accumulation of combined point mutations, which can increase the risk of TKI-resistance formation.
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): BCR-ABL, Resistance, Tyrosine kinase inhibitor
Abstract: PB1894
Type: Publication Only
Background
One of the most controversial causes of TKI resistance in patients with CML is the pathogenic BCR- ABL del. c.1086-1270 transcript. This transcript was first described in 2008 in TKI resistant patients (Curvo et al., 2008). BCR-ABL del. c.1086-1270 transcript encodes truncated fusion protein BCR-ABL p.R362fs*21 which is an alternative splicing product. It has been shown that such truncated protein is unlikely to have tyrosine kinase activity because of the disfunction of the ATP-binding site (Meggyesi et al., 2012). However, we suggest that pathogenic effect of BCR-ABL p.R362fs*21 expression is possible due to the dimerization with a typical protein BCR-ABL p210. Such mechanism has already been proved for the splice variant of serine-threonine kinase BRAF (Poulikakos etal., 2011).
Aims
To evaluate the pathogenic effect of BCR-ABL del. c.1086-1270 (p.R362fs*21) on TKI resistance formation in Russian patients with CML.
Methods
33 male and 50 female CML patients (age 24-80) with BCR-ABL transcript level > 0.1% were included in the study. BCR-ABL exon 7 deletion was analyzed with two round (nested) PCR followed by Sanger sequencing. Initial screening for deletions was performed by means of fragment analysis (Applied Biosystems 3130).
Results
Fragment analysis detected BCR-ABL del. c.1086-1270 transcript in 32 patients (38%). The ratio of TKI-sensitive to TKI-resistant among these 32 patients was approximately 47% (15 patients) to 53% (17 patients) respectively. Direct sequencing showed the tumor clone, which contains the BCR-ABL1 del.c.1086-1270 always paired with a normal clone of BCR-ABL p210, in all TKI-resistant patients. This fact was verified by the results of agarose gel electrophoresis. We suggest, that these results show an indirect evidence of the dimerization of the protein BCR-ABL del. c.1086-1270 with chimeric protein Bcr-Abl p210. Furthermore, the deletion clone of each TKI-resistant patient always has a point mutation (F317V, F317L, E282Q, M351T, T315I) which is unlikely for the normal clone (Figure 1). This fact contradicts the alternative splicing theory.

Conclusion
Our results show a definite correlation between the presence of the deletion and TKI-resistance. BCR- ABL1 del.c.1086-1270 is likely to play a trigger-role in TKI-resistance formation: clones with the exon 7 deletion are extremely prone to the accumulation of combined point mutations, which can increase the risk of TKI-resistance formation.
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): BCR-ABL, Resistance, Tyrosine kinase inhibitor


