
Contributions
Abstract: PB1891
Type: Publication Only
Background
Today the gold standard analyses to characterize patients affected by chronic myeloid leukemia (CML) require bone marrow samples, however peripheral blood may be used to monitor the molecular response during tyrosine kinase inhibitors (TKIs) treatment. Recent studies have shown that saliva could be an alternative substrate for biomarker detection in many diseases. To date, there are no preemptive studies that investigate the salivary proteomic profile to identify non-responders to TKI therapy in CML.
Aims
The aim is to identify putative response biomarkers in the saliva of CML patients.
Methods
We investigated the salivary expression profile of 176 proteins at two different time points in 2 groups of patients with CML. The first group was represented by 4 patients in stable deep molecular response during TKIs treatment, while the second one included 4 sex and age-matched TKI-failure cases. Analyses were performed at baseline and after 6 months of treatment with TKIs.
Proteomic analysis was performed by using albumin- and IgG-depleted saliva samples through a nano-HPLC system coupled with a TripleTOFTM 5600 mass spectrometer. Statistical comparative analysis was performed using PeakViewTM Software with SWATHTM Acquisition MicroApp 2.0 and MarkerViewTM.
Results
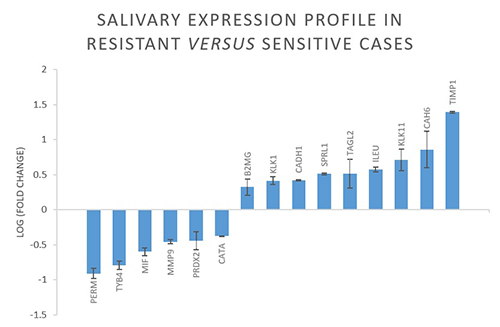
Overall, 64 proteins resulted differently expressed in resistant versus sensitive cases. As shown in the image, myeloperoxidase (PERM), thymosine beta-4 (TYB4), matrix metalloproteinase-9 (MMP9), peroxiredoxin-2 (PRDX2), catalase (CATA) and macrophage migration inhibitory factor (MIF) were down-regulated, while MMP-9-inhibitor (TIMP1), SPARC-like protein-1 (SPRL1), transgelin-2 (TAGL2), leucocyte elastase inhibitor (ILEU), carbonic anhydrase-6 (CAH6), kallikrein-1 (KLK1) and -11 (KLK11), cadherin-1 (CADH1) and beta-2-microglobulin (B2MG) were over-expressed in resistant patients compared to sensitive ones.
Conclusion
Our results show that in resistant patients, proteins implicated in hypoxia of the niche are over-expressed. This may be implied in the induction of resistance to TKIs because of the lower BCR-ABL1 expression. In details, these proteins are involved in differentiation of the osteoblasts (PRDX2, MIF), which are main cellular components of the niche; in myeloid differentiation (TYB4, ILEU, PERM), which is a sign of response to TKIs therapy; in fibrosis of the niche (MMP-9, TIMP1), thus preventing the oxygenation of the leukemic stem cell (LSC) and consequently its protein expression; in tumor-induced angiogenesis (MIF, TAGL2); in LSC quiescence (CATA); in components of the niche microenviroment (SPRL1, CAH6); in factors that support the adhesion of LSC to the stroma (TAGL2, CADH1). In addition, some of these proteins have been already described as prognostic factors in solid tumors (MIF, KLK1, KLK11) and leukemias (B2MG, MIF, CADH1), while CADH1 seems to be correlated to a better response to imatinib in CML patients. Our data show that proteins traditionally identified in peripheral blood only, can be detected also in the saliva, thus indicating that use of this compartment for characterization of CML patients may be feasible. Moreover, the salivary expression profile in resistant versus sensitive cases is significantly different and coherent with their response to TKIs treatment, thus suggesting salivary proteomic profile as a possible method to identify the occurrence of resistance to TKIs. However, further studies are needed to indicate saliva as a valid alternative to peripheral blood and bone marrow to monitor response to TKIs treatment in CML patients.
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): Chronic myeloid leukemia, Monitor, Resistance
Abstract: PB1891
Type: Publication Only
Background
Today the gold standard analyses to characterize patients affected by chronic myeloid leukemia (CML) require bone marrow samples, however peripheral blood may be used to monitor the molecular response during tyrosine kinase inhibitors (TKIs) treatment. Recent studies have shown that saliva could be an alternative substrate for biomarker detection in many diseases. To date, there are no preemptive studies that investigate the salivary proteomic profile to identify non-responders to TKI therapy in CML.
Aims
The aim is to identify putative response biomarkers in the saliva of CML patients.
Methods
We investigated the salivary expression profile of 176 proteins at two different time points in 2 groups of patients with CML. The first group was represented by 4 patients in stable deep molecular response during TKIs treatment, while the second one included 4 sex and age-matched TKI-failure cases. Analyses were performed at baseline and after 6 months of treatment with TKIs.
Proteomic analysis was performed by using albumin- and IgG-depleted saliva samples through a nano-HPLC system coupled with a TripleTOFTM 5600 mass spectrometer. Statistical comparative analysis was performed using PeakViewTM Software with SWATHTM Acquisition MicroApp 2.0 and MarkerViewTM.
Results
Overall, 64 proteins resulted differently expressed in resistant versus sensitive cases. As shown in the image, myeloperoxidase (PERM), thymosine beta-4 (TYB4), matrix metalloproteinase-9 (MMP9), peroxiredoxin-2 (PRDX2), catalase (CATA) and macrophage migration inhibitory factor (MIF) were down-regulated, while MMP-9-inhibitor (TIMP1), SPARC-like protein-1 (SPRL1), transgelin-2 (TAGL2), leucocyte elastase inhibitor (ILEU), carbonic anhydrase-6 (CAH6), kallikrein-1 (KLK1) and -11 (KLK11), cadherin-1 (CADH1) and beta-2-microglobulin (B2MG) were over-expressed in resistant patients compared to sensitive ones.

Conclusion
Our results show that in resistant patients, proteins implicated in hypoxia of the niche are over-expressed. This may be implied in the induction of resistance to TKIs because of the lower BCR-ABL1 expression. In details, these proteins are involved in differentiation of the osteoblasts (PRDX2, MIF), which are main cellular components of the niche; in myeloid differentiation (TYB4, ILEU, PERM), which is a sign of response to TKIs therapy; in fibrosis of the niche (MMP-9, TIMP1), thus preventing the oxygenation of the leukemic stem cell (LSC) and consequently its protein expression; in tumor-induced angiogenesis (MIF, TAGL2); in LSC quiescence (CATA); in components of the niche microenviroment (SPRL1, CAH6); in factors that support the adhesion of LSC to the stroma (TAGL2, CADH1). In addition, some of these proteins have been already described as prognostic factors in solid tumors (MIF, KLK1, KLK11) and leukemias (B2MG, MIF, CADH1), while CADH1 seems to be correlated to a better response to imatinib in CML patients. Our data show that proteins traditionally identified in peripheral blood only, can be detected also in the saliva, thus indicating that use of this compartment for characterization of CML patients may be feasible. Moreover, the salivary expression profile in resistant versus sensitive cases is significantly different and coherent with their response to TKIs treatment, thus suggesting salivary proteomic profile as a possible method to identify the occurrence of resistance to TKIs. However, further studies are needed to indicate saliva as a valid alternative to peripheral blood and bone marrow to monitor response to TKIs treatment in CML patients.
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): Chronic myeloid leukemia, Monitor, Resistance


