
Contributions
Abstract: PB1896
Type: Publication Only
Background
After the introduction in the clinical practice of tyrosine kinase inhibitors (TKIs),the overall survival of CML patients is really improved, but several mechanisms of resistance have been reported. In addition to BCR-ABL1-related mechanisms (amplification, ABL1 mutations), the persistence of the leukemic stem cell (LSC) in the bone marrow niche is a very relevant problem. The hypoxia and the presence of immunosuppressive cells in the microenvironment are well-known mechanisms that sustain the LSC; nevertheless, an increasing interest is also put today into the WNT/Beta-catenin pathway, that is necessary to self-renewal of normal cells, but its deregulation also causes leukemogenesis and progression in several types of cancers (Zhao, 2007).
Aims
we decided to assess the expression of 86 genes of the beta-catenin/WNT pathway at diagnosis and after 6 months of treatment with TKIs in a cohort of 10 patients with different responses to treatment.
Methods
Buffy coats obtained from peripheral blood samples of 10 patients (7receiving imatinib, 2 nilotinib, and 1 dasatinib) have been used for the total RNA extraction. In addition to the quantification of the BCR-ABL1/ABL1 ratio %IS, we used RT-q PCR to measure the expression of 86 genes from the WNT pathway (PrimePCR pathway kit, Biorad, Milan, Italy) at diagnosis and after 6 months of therapy. Expression values were calculated by the Vandesompele method using four housekeeping genes. Data have been analyzed by the “Gene Study” PrimePCR analysis software (Biorad).
Results
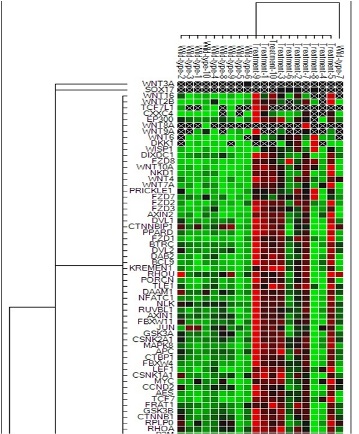
Five patients achieved an optimal response and five were no responders, according to the European Leukemia Network guidelines. Interestingly, after 6 months of treatment, we observed a de-regulation of 36 genes. Down-expression occurred in 14% of genes, while 79% of genes were up-regulated. When we compared the change of expression with the quality of response to TKIs, a differential expression between patients with or without an optimal response was observed: in the cases without optimal response, we found as up-regulated: 1) FZD7, already known to be responsible for the protection of leukemic CML cell, proliferation and drug resistance in K562 cells; 2) WNT6, that predicts unfavorable survival in solid cancer and whose expression is inversely correlated to the responseto ECF (Epi, cisplatin, 5-fluorouracil) chemotherapy in human gastric cancer cells; 3)WISP1, with anti-apoptotic activity, associated to poor prognosis and advanced stage in glioblastoma. On the other hand, the most frequently down-regulated gene was CSNK1A1, whose aploinsufficiency has been shown to result in a more probable transformation of myelodysplastic syndrome (MDS) in acute myeloid leukemia (AML) and to induce proliferation, invasion and metastasis in multiple myeloma (MM), lymphoma (DLBCL) and AML.
Conclusion
With these experiments of gene expression profiling we demonstrated, although in a small group of CML patients, that the beta-catenin /WNT pathway might be relevant to condition the response to TKIs. Obviously, the analysis of a larger number of patients will improve the biological suggestions coming from these preliminary data
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): Gene regulation, Tyrosine kinase inhibitor, Wnt
Abstract: PB1896
Type: Publication Only
Background
After the introduction in the clinical practice of tyrosine kinase inhibitors (TKIs),the overall survival of CML patients is really improved, but several mechanisms of resistance have been reported. In addition to BCR-ABL1-related mechanisms (amplification, ABL1 mutations), the persistence of the leukemic stem cell (LSC) in the bone marrow niche is a very relevant problem. The hypoxia and the presence of immunosuppressive cells in the microenvironment are well-known mechanisms that sustain the LSC; nevertheless, an increasing interest is also put today into the WNT/Beta-catenin pathway, that is necessary to self-renewal of normal cells, but its deregulation also causes leukemogenesis and progression in several types of cancers (Zhao, 2007).
Aims
we decided to assess the expression of 86 genes of the beta-catenin/WNT pathway at diagnosis and after 6 months of treatment with TKIs in a cohort of 10 patients with different responses to treatment.
Methods
Buffy coats obtained from peripheral blood samples of 10 patients (7receiving imatinib, 2 nilotinib, and 1 dasatinib) have been used for the total RNA extraction. In addition to the quantification of the BCR-ABL1/ABL1 ratio %IS, we used RT-q PCR to measure the expression of 86 genes from the WNT pathway (PrimePCR pathway kit, Biorad, Milan, Italy) at diagnosis and after 6 months of therapy. Expression values were calculated by the Vandesompele method using four housekeeping genes. Data have been analyzed by the “Gene Study” PrimePCR analysis software (Biorad).
Results
Five patients achieved an optimal response and five were no responders, according to the European Leukemia Network guidelines. Interestingly, after 6 months of treatment, we observed a de-regulation of 36 genes. Down-expression occurred in 14% of genes, while 79% of genes were up-regulated. When we compared the change of expression with the quality of response to TKIs, a differential expression between patients with or without an optimal response was observed: in the cases without optimal response, we found as up-regulated: 1) FZD7, already known to be responsible for the protection of leukemic CML cell, proliferation and drug resistance in K562 cells; 2) WNT6, that predicts unfavorable survival in solid cancer and whose expression is inversely correlated to the responseto ECF (Epi, cisplatin, 5-fluorouracil) chemotherapy in human gastric cancer cells; 3)WISP1, with anti-apoptotic activity, associated to poor prognosis and advanced stage in glioblastoma. On the other hand, the most frequently down-regulated gene was CSNK1A1, whose aploinsufficiency has been shown to result in a more probable transformation of myelodysplastic syndrome (MDS) in acute myeloid leukemia (AML) and to induce proliferation, invasion and metastasis in multiple myeloma (MM), lymphoma (DLBCL) and AML.

Conclusion
With these experiments of gene expression profiling we demonstrated, although in a small group of CML patients, that the beta-catenin /WNT pathway might be relevant to condition the response to TKIs. Obviously, the analysis of a larger number of patients will improve the biological suggestions coming from these preliminary data
Session topic: 7. Chronic myeloid leukemia – Biology & Translational Research
Keyword(s): Gene regulation, Tyrosine kinase inhibitor, Wnt


