
Contributions
Abstract: PB1691
Type: Publication Only
Background
Acute Myeloid Leukemia (AML) cases presenting intermediate-prognostic cytogenetic features account for about 40% of adult AMLs and display heterogeneous clinical outcomes.
Over the past 2 decades an increasing number of molecular lesions, having prognostic and therapeutic significance, have been described in AML. Mounting evidence is showing that deep genomic characterization might provide an important tool to predict pathology progression.
In line with these findings, two recent works (Grimwade et al., 2015 and Pappaemmanuil et al.,2016) proposed a genomic classification of AML that relies upon the study of a number of recurrent mutations.
Aims
Validation of a specific gene panel, having prognostic and therapeutic relevance, on AML patients (preliminary results).
Methods
Taking advantage of Next Generation Sequencing (NGS) Illumina MiSeq platform, we performed deep sequencing analysis of the following genes: ABL (exons 4-9), ASXL1 (exons 9,11,12,14), BRAF (exon 15), CALR (exon 9), CBL (exons 8,9), CEBPA (all gene), CSF3R (all gene), DNMT3A (all gene), ETV6 (all gene), EZH2 (all gene), FLTR3 (exons 13-15,20), HRAS (exons 2,3), IDH1 (exon 4), IDH2 (exon 4), JAK2 (all gene), KIT (exons 2,8-11,13,17,18), KRAS (exons 2,3), MPL (exon 10), NPM1 (exons 10,11), NRAS (exons 2,3), PTPN11 (exons 3,7-13), RUNX1 (all gene), SETBP1 (exon 4), SF3B1 (exons 10-16), SRSF2 (exon 1), TET2 (all gene), TP53 (exons 2-11), U2AF1 (exons 2,6), WT1 (exons 6-10), ZRSR2 (all gene).
In this study, AML cases at onset, that presented intermediate-prognostic cytogenetic risk were considered. We analyzed a total of 11 samples, 5 of which had normal karyotype, 5 patients harboured intermediate cytogenetic alterations, 1 case had complex karyotype.
Results
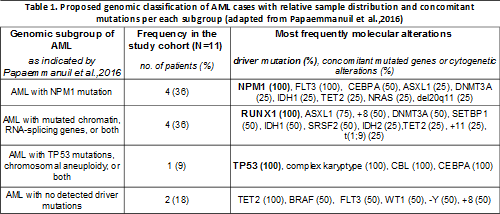
From our analysis we were able to collect all analyzed AMLs into molecularly-defined subsets, according to what reported by Grimwade and Pappaemmanuil and colleagues. In particular, we observed 36% of cases harbouring NPM1 as driver-mutation, of which 100% displayed co-occurrence of FLT3 mutation. On the other hand, 36% of cases were RUNX1-mutated. In this latter group, 75% of cases associated with lesions in ASXL1 gene and 50% with additional chromosome 8. Finally, 18% of cases did not meet criteria with classes presenting NPM1 or RUNX1 driver mutations, whereas the sample with complex karyotype displayed simultaneous TP53 mutation (see table 1).
Importantly, we noticed that all AML samples, apart from the one with complex karyotype, had molecular alterations on epigenetic modifiers, mainly IDH1-2, DNMT3A or, alternatively, TET2, that displayed Variant Allele Frequencies (VAF) equal to or higher than 0.5, suggesting that aberrant epigenetic pathways affect the majority of blast cells, therefore representing an early event in AML pathogenesis.
Conclusion
Although the combinatorial effect of different molecular lesions remains to be uncovered, our data show that the emerging genomic classification system might represent a reproducible method for AML characterization based on molecular approaches.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Molecular markers, mutation analysis
Abstract: PB1691
Type: Publication Only
Background
Acute Myeloid Leukemia (AML) cases presenting intermediate-prognostic cytogenetic features account for about 40% of adult AMLs and display heterogeneous clinical outcomes.
Over the past 2 decades an increasing number of molecular lesions, having prognostic and therapeutic significance, have been described in AML. Mounting evidence is showing that deep genomic characterization might provide an important tool to predict pathology progression.
In line with these findings, two recent works (Grimwade et al., 2015 and Pappaemmanuil et al.,2016) proposed a genomic classification of AML that relies upon the study of a number of recurrent mutations.
Aims
Validation of a specific gene panel, having prognostic and therapeutic relevance, on AML patients (preliminary results).
Methods
Taking advantage of Next Generation Sequencing (NGS) Illumina MiSeq platform, we performed deep sequencing analysis of the following genes: ABL (exons 4-9), ASXL1 (exons 9,11,12,14), BRAF (exon 15), CALR (exon 9), CBL (exons 8,9), CEBPA (all gene), CSF3R (all gene), DNMT3A (all gene), ETV6 (all gene), EZH2 (all gene), FLTR3 (exons 13-15,20), HRAS (exons 2,3), IDH1 (exon 4), IDH2 (exon 4), JAK2 (all gene), KIT (exons 2,8-11,13,17,18), KRAS (exons 2,3), MPL (exon 10), NPM1 (exons 10,11), NRAS (exons 2,3), PTPN11 (exons 3,7-13), RUNX1 (all gene), SETBP1 (exon 4), SF3B1 (exons 10-16), SRSF2 (exon 1), TET2 (all gene), TP53 (exons 2-11), U2AF1 (exons 2,6), WT1 (exons 6-10), ZRSR2 (all gene).
In this study, AML cases at onset, that presented intermediate-prognostic cytogenetic risk were considered. We analyzed a total of 11 samples, 5 of which had normal karyotype, 5 patients harboured intermediate cytogenetic alterations, 1 case had complex karyotype.
Results
From our analysis we were able to collect all analyzed AMLs into molecularly-defined subsets, according to what reported by Grimwade and Pappaemmanuil and colleagues. In particular, we observed 36% of cases harbouring NPM1 as driver-mutation, of which 100% displayed co-occurrence of FLT3 mutation. On the other hand, 36% of cases were RUNX1-mutated. In this latter group, 75% of cases associated with lesions in ASXL1 gene and 50% with additional chromosome 8. Finally, 18% of cases did not meet criteria with classes presenting NPM1 or RUNX1 driver mutations, whereas the sample with complex karyotype displayed simultaneous TP53 mutation (see table 1).
Importantly, we noticed that all AML samples, apart from the one with complex karyotype, had molecular alterations on epigenetic modifiers, mainly IDH1-2, DNMT3A or, alternatively, TET2, that displayed Variant Allele Frequencies (VAF) equal to or higher than 0.5, suggesting that aberrant epigenetic pathways affect the majority of blast cells, therefore representing an early event in AML pathogenesis.

Conclusion
Although the combinatorial effect of different molecular lesions remains to be uncovered, our data show that the emerging genomic classification system might represent a reproducible method for AML characterization based on molecular approaches.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Molecular markers, mutation analysis


