
Contributions
Abstract: PB1682
Type: Publication Only
Background
The use of ATRA combined with chemotherapy (CHT) and/or arsenic trioxide (ATO) can achieve long-term remission in the vast majority of patients with newly diagnosed APL. Despite this therapeutic success, 5-10% patients still relapse after modern treatment. Up to 30% of relapsed/refractory patients harbor point mutations within the ATO-binding domain (B2) of the PML moiety of the PML/RARA hybrid. These alterations affect the binding of ATO to the oncoprotein and thus impair PML/RARA degradation and clinical response to this agent. The most common mutation detected in APL refractory to ATO is the A216V, which is located within the PML-B2 domain.
Aims
In the present study, we developed and tested a droplet digital PCR (ddPCR) assay for the sensitive detection of PML-A216V mutation in relapsed APL patients as a tool to early predict ATO-resistance.
Methods
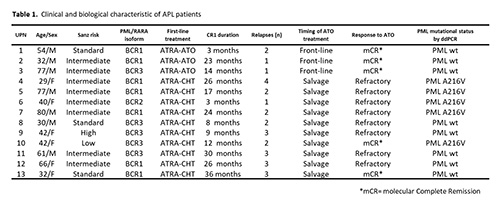
A total of 13 patients who relapsed after ATO treatment were analyzed. Table 1 shows the main patient characteristics at the time of APL diagnosis. Initial treatment included ATRA/CHT in 10 and ATO/ATRA in 3 cases. Eleven patients in the series underwent multiple relapses. Mutational analysis was performed at the time of first relapse in the 3 patients treated with front-line ATO and in ≥2nd relapse in patients receiving ATO as salvage treatment. Mutational analysis of PML-A216V by ddPCR assay (BioRad), was performed in duplicate, using 50 ng of DNA for each sample. The limit of detection (LOD) of ddPCR assay was determined by diluting mutant DNA in the DNA sample derived from a healthy donor at the following ratios: 1:10, 1:100, 1:250 and 1:500. The threshold for positive amplification was established within each reaction based on the results of PML-A216V negative template controls. The ddPCR assay was initially performed on DNA samples collected at the time of relapse where a high PML/RARA copy number had been detected by conventional RQ-PCR. We then investigated the mutation dynamics by backtracking the identified mutation in samples collected for routine PML/RARA monitoring prior to relapse. To confirm and compare the results obtained by ddPCR, all positive samples were also analyzed by Sanger sequencing.
Results
The ddPCR test showed high reproducibility and sensitivity and was able to detect up to 0.4% PML-A216V-mutant allele fraction. After assessing the false positive rate (FPR), samples were considered positive if the mutation rate had ≥3 positive droplets above the threshold of the negative template controls. The A216V mutation was detected by ddPCR in 5/13 patients (38%) who relapsed after ATO. Sanger sequencing allowed to identify the PML-A216V mutation in 4/5 cases. The ddPCR assay carried out in follow-up DNAs of mutated patients (total of 44 samples, median 3 per patient, range:1-22), revealed the presence of PML-A216V mutation in 17 samples. Of these, 3 were collected in overt relapse while 14 were taken at molecular relapse in patients showing low PML/RARA transcript levels. Sanger sequencing confirmed the mutation in only 4/17 samples. In 3 mutated patients for whom several sequential samples were available, a positive-ddPCR test anticipated a positive Sanger sequencing result by 3, 4 and 24 months, respectively.
Conclusion
Our data show that a ddPCR assay can be efficiently employed in the screening of PML-A216V mutation in APL and is able to identify mutant cases earlier in the disease course as compared to conventional sequencing. This sensitive method may help to identify ATO-resistant APL patients who are candidate to alternative treatment strategies.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): Acute Promyelocytic Leukemia, Arsenic trioxide, PML
Abstract: PB1682
Type: Publication Only
Background
The use of ATRA combined with chemotherapy (CHT) and/or arsenic trioxide (ATO) can achieve long-term remission in the vast majority of patients with newly diagnosed APL. Despite this therapeutic success, 5-10% patients still relapse after modern treatment. Up to 30% of relapsed/refractory patients harbor point mutations within the ATO-binding domain (B2) of the PML moiety of the PML/RARA hybrid. These alterations affect the binding of ATO to the oncoprotein and thus impair PML/RARA degradation and clinical response to this agent. The most common mutation detected in APL refractory to ATO is the A216V, which is located within the PML-B2 domain.
Aims
In the present study, we developed and tested a droplet digital PCR (ddPCR) assay for the sensitive detection of PML-A216V mutation in relapsed APL patients as a tool to early predict ATO-resistance.
Methods
A total of 13 patients who relapsed after ATO treatment were analyzed. Table 1 shows the main patient characteristics at the time of APL diagnosis. Initial treatment included ATRA/CHT in 10 and ATO/ATRA in 3 cases. Eleven patients in the series underwent multiple relapses. Mutational analysis was performed at the time of first relapse in the 3 patients treated with front-line ATO and in ≥2nd relapse in patients receiving ATO as salvage treatment. Mutational analysis of PML-A216V by ddPCR assay (BioRad), was performed in duplicate, using 50 ng of DNA for each sample. The limit of detection (LOD) of ddPCR assay was determined by diluting mutant DNA in the DNA sample derived from a healthy donor at the following ratios: 1:10, 1:100, 1:250 and 1:500. The threshold for positive amplification was established within each reaction based on the results of PML-A216V negative template controls. The ddPCR assay was initially performed on DNA samples collected at the time of relapse where a high PML/RARA copy number had been detected by conventional RQ-PCR. We then investigated the mutation dynamics by backtracking the identified mutation in samples collected for routine PML/RARA monitoring prior to relapse. To confirm and compare the results obtained by ddPCR, all positive samples were also analyzed by Sanger sequencing.
Results
The ddPCR test showed high reproducibility and sensitivity and was able to detect up to 0.4% PML-A216V-mutant allele fraction. After assessing the false positive rate (FPR), samples were considered positive if the mutation rate had ≥3 positive droplets above the threshold of the negative template controls. The A216V mutation was detected by ddPCR in 5/13 patients (38%) who relapsed after ATO. Sanger sequencing allowed to identify the PML-A216V mutation in 4/5 cases. The ddPCR assay carried out in follow-up DNAs of mutated patients (total of 44 samples, median 3 per patient, range:1-22), revealed the presence of PML-A216V mutation in 17 samples. Of these, 3 were collected in overt relapse while 14 were taken at molecular relapse in patients showing low PML/RARA transcript levels. Sanger sequencing confirmed the mutation in only 4/17 samples. In 3 mutated patients for whom several sequential samples were available, a positive-ddPCR test anticipated a positive Sanger sequencing result by 3, 4 and 24 months, respectively.

Conclusion
Our data show that a ddPCR assay can be efficiently employed in the screening of PML-A216V mutation in APL and is able to identify mutant cases earlier in the disease course as compared to conventional sequencing. This sensitive method may help to identify ATO-resistant APL patients who are candidate to alternative treatment strategies.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): Acute Promyelocytic Leukemia, Arsenic trioxide, PML


