
Contributions
Abstract: PB1701
Type: Publication Only
Background
Whole-genome sequencing has revealed a central role of epigenetic modulators in acute myeloid leukemia (AML) pathogenesis. Targeted therapy has also focused on epigenetics as seen by the use of IDH and BET-inhibitors. We believe that establishing the expression profile of epigenetic modulators in AML may help in the understanding of AML genesis, biology, evolution and treatment.
Aims
We wanted to get a better insight regarding the expression profile of epigenetic modulators in AML of intermediate risk by studying expression levels of EZH2, ASXL1, TET2, BRD4, c-myc and Bcl-2 in a consecutive series of AML patients at diagnosis and after induction and its association with clinical evolution.
Methods
Our series consisted of 120 intermediate-risk AML patients with a mean age of 56.04 years (range 15-79 years), diagnosed and homogenously treated between 2005 and 2017 at the Hospital Universitario de Gran Canaria Dr. Negrín with a median follow up of 14 months. Gene expression analyses were carried out by real time PCR in a LightCycler 480 Instrument II (Roche) using ABL as a control gene. Results were normalized with a cDNA pool from the bone marrow of 10 healthy donors, which was introduced as an internal control in each experiment. For statistical analyses, SPSS (v.15.0) software was used.
Results
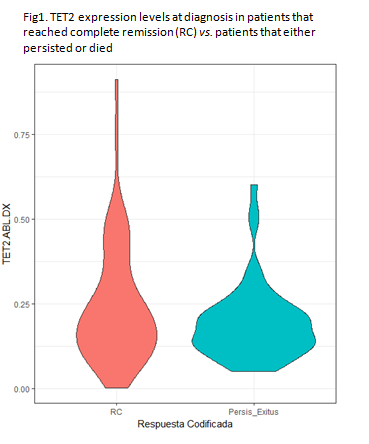
ASXL1 levels were positively associated with EZH2 (Spearman's coefficient, SC=0.4, p<0.001). BRD4 expression was strongly correlated with ASXL1 (SC=0.56, p<0.001) and EZH2 (SC=0.31, p<0.001) and with c-myc expression (SC= 0.25, p=0.01). On the other hand, TET2 expression showed a positive relationship with BRD4 (SC=0.23, p=0.01), EZH2 (SC=0.24, p=0.01) and ASXL1 (SC=0.2, p=0.01). As shown in Fig.1 TET2 levels at diagnosis were marginally significantly lower in those cases that persisted or died compared to those that reached complete remission after induction (mean 0.19 vs 0.24, p=0.06). Measurement of BRD4 levels after induction showed that patients that displayed a reduction of more than 95% compared to levels at diagnosis exhibited worse overall survival (median overall survival, OS, 14 months vs. 28 months, p=0.046).
Conclusion
The positive association observed between EZH2 and ASXL1 agrees with the fact that both cooperate in adding the epigenetic repressive H3K27 mark. In general, it seems that expression levels of genes that behave as tumor suppressor genes in AML, such us TET2, ASXL2 and EZH2, correlate well with BRD4 expression. BRD4 and c-myc levels were associated in accordance with what has previously been described, since BRD4 binds c-myc superenhancer regions, activating its expression. In contrast, lower levels of TET2 at diagnosis and a reduction of BRD4 levels after induction compared to diagnosis seemed to be related to worse prognosis. Further studies with a larger series are needed to confirm these preliminary results.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Epigenetic, Expression profiling
Abstract: PB1701
Type: Publication Only
Background
Whole-genome sequencing has revealed a central role of epigenetic modulators in acute myeloid leukemia (AML) pathogenesis. Targeted therapy has also focused on epigenetics as seen by the use of IDH and BET-inhibitors. We believe that establishing the expression profile of epigenetic modulators in AML may help in the understanding of AML genesis, biology, evolution and treatment.
Aims
We wanted to get a better insight regarding the expression profile of epigenetic modulators in AML of intermediate risk by studying expression levels of EZH2, ASXL1, TET2, BRD4, c-myc and Bcl-2 in a consecutive series of AML patients at diagnosis and after induction and its association with clinical evolution.
Methods
Our series consisted of 120 intermediate-risk AML patients with a mean age of 56.04 years (range 15-79 years), diagnosed and homogenously treated between 2005 and 2017 at the Hospital Universitario de Gran Canaria Dr. Negrín with a median follow up of 14 months. Gene expression analyses were carried out by real time PCR in a LightCycler 480 Instrument II (Roche) using ABL as a control gene. Results were normalized with a cDNA pool from the bone marrow of 10 healthy donors, which was introduced as an internal control in each experiment. For statistical analyses, SPSS (v.15.0) software was used.
Results
ASXL1 levels were positively associated with EZH2 (Spearman's coefficient, SC=0.4, p<0.001). BRD4 expression was strongly correlated with ASXL1 (SC=0.56, p<0.001) and EZH2 (SC=0.31, p<0.001) and with c-myc expression (SC= 0.25, p=0.01). On the other hand, TET2 expression showed a positive relationship with BRD4 (SC=0.23, p=0.01), EZH2 (SC=0.24, p=0.01) and ASXL1 (SC=0.2, p=0.01). As shown in Fig.1 TET2 levels at diagnosis were marginally significantly lower in those cases that persisted or died compared to those that reached complete remission after induction (mean 0.19 vs 0.24, p=0.06). Measurement of BRD4 levels after induction showed that patients that displayed a reduction of more than 95% compared to levels at diagnosis exhibited worse overall survival (median overall survival, OS, 14 months vs. 28 months, p=0.046).

Conclusion
The positive association observed between EZH2 and ASXL1 agrees with the fact that both cooperate in adding the epigenetic repressive H3K27 mark. In general, it seems that expression levels of genes that behave as tumor suppressor genes in AML, such us TET2, ASXL2 and EZH2, correlate well with BRD4 expression. BRD4 and c-myc levels were associated in accordance with what has previously been described, since BRD4 binds c-myc superenhancer regions, activating its expression. In contrast, lower levels of TET2 at diagnosis and a reduction of BRD4 levels after induction compared to diagnosis seemed to be related to worse prognosis. Further studies with a larger series are needed to confirm these preliminary results.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Epigenetic, Expression profiling


