
Contributions
Abstract: PB1692
Type: Publication Only
Background
Acute myeloid leukemia (AML) is the most frequently diagnosed type of acute leukemia in adults, with a dismal 5-year relative survival of only 25.9%. Efforts to improve prognosis focus on elucidating the mechanism behind AML development, to improve risk classification using novel biomarker and establishing new means for personalized treatment. Non-coding parts of the transcriptome, such as long non-coding RNAs (lncRNAs) exhibit crucial regulating functions and their dysfunction is often associated with cancer. However, the functional role of lncRNAs in the development and progression of AML is so far poorly understood.
Aims
In this study, we aimed to identify long non-coding RNAs (lncRNAs), which are differentially expressed in AML and study their role on leukemogenesis. We hypothesized that these non-coding transcripts could potentially play important biological roles in the development or progression of AML, serve as novel prognostic or predictive biomarkers or could indicate new treatment targets in AML.
Methods
We performed deep RNA-sequencing in a subset of AML samples and normal CD34+ bone marrow cells and identified, among others, lncRNA XLOC_091701 (internal reference), which is upregulated in AML. TSS peaks of XLOC_091701 were confirmed using FANTOM CAGE data (RIKEN) and XLOC_091701 expression was validated in ten myeloid cell lines using RT-qPCR. Further, XLOC_091701 expression was validated using RNA expression data from our AML patient cohort (n=325, ClinSeq cohort) and confirmed by The Cancer Genome Atlas (TCGA)-LAML cohort (n=151). Next, multivariate cox regression analysis and clinical correlation analysis was done to identify significant correlations with clinical parameters and survival using available clinical data on intensively treated AML patients (n=268, ClinSeq-AML cohort).
Results
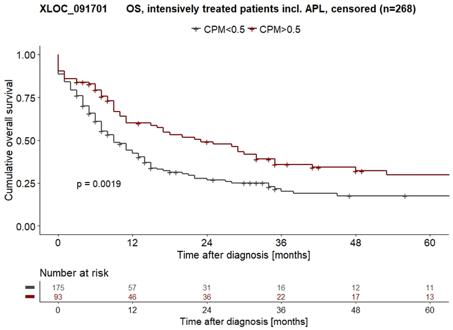
We found XLOC_091701 to be exclusively expressed in certain myeloid cell lines, especially in HL60 and U937 cells, and expressed in about a third of all tested AML patients in both ClinSeq-AML cohort (n=268) and TCGA-LAML cohort (n=117, cut-off: CPM>0.5). Next, multivariate cox regression revealed significant superior survival in patients with XLOC_091701 expression independent of other variates known for good prognosis (n=268, ClinSeq-AML cohort, p=0.002) and moreover associates with formerly used AML FAB classification M2 and M3 (APL) along with low genetic and low cytogenetic risk groups.
Conclusion
Here, we describe a long non-coding RNA transcript (XLOC_091701) whose expression is associated with better overall survival in AML patients. Our findings suggest that XLOC_091701 plays a biological role in AML and might serve as prognostic biomarker in clinical settings. Currently, we are creating a KO-model to characterize XLOC_091701 functionally to explore the regulatory mechanisms behind the observed favorable prognosis.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Molecular markers, Outcome
Abstract: PB1692
Type: Publication Only
Background
Acute myeloid leukemia (AML) is the most frequently diagnosed type of acute leukemia in adults, with a dismal 5-year relative survival of only 25.9%. Efforts to improve prognosis focus on elucidating the mechanism behind AML development, to improve risk classification using novel biomarker and establishing new means for personalized treatment. Non-coding parts of the transcriptome, such as long non-coding RNAs (lncRNAs) exhibit crucial regulating functions and their dysfunction is often associated with cancer. However, the functional role of lncRNAs in the development and progression of AML is so far poorly understood.
Aims
In this study, we aimed to identify long non-coding RNAs (lncRNAs), which are differentially expressed in AML and study their role on leukemogenesis. We hypothesized that these non-coding transcripts could potentially play important biological roles in the development or progression of AML, serve as novel prognostic or predictive biomarkers or could indicate new treatment targets in AML.
Methods
We performed deep RNA-sequencing in a subset of AML samples and normal CD34+ bone marrow cells and identified, among others, lncRNA XLOC_091701 (internal reference), which is upregulated in AML. TSS peaks of XLOC_091701 were confirmed using FANTOM CAGE data (RIKEN) and XLOC_091701 expression was validated in ten myeloid cell lines using RT-qPCR. Further, XLOC_091701 expression was validated using RNA expression data from our AML patient cohort (n=325, ClinSeq cohort) and confirmed by The Cancer Genome Atlas (TCGA)-LAML cohort (n=151). Next, multivariate cox regression analysis and clinical correlation analysis was done to identify significant correlations with clinical parameters and survival using available clinical data on intensively treated AML patients (n=268, ClinSeq-AML cohort).
Results
We found XLOC_091701 to be exclusively expressed in certain myeloid cell lines, especially in HL60 and U937 cells, and expressed in about a third of all tested AML patients in both ClinSeq-AML cohort (n=268) and TCGA-LAML cohort (n=117, cut-off: CPM>0.5). Next, multivariate cox regression revealed significant superior survival in patients with XLOC_091701 expression independent of other variates known for good prognosis (n=268, ClinSeq-AML cohort, p=0.002) and moreover associates with formerly used AML FAB classification M2 and M3 (APL) along with low genetic and low cytogenetic risk groups.

Conclusion
Here, we describe a long non-coding RNA transcript (XLOC_091701) whose expression is associated with better overall survival in AML patients. Our findings suggest that XLOC_091701 plays a biological role in AML and might serve as prognostic biomarker in clinical settings. Currently, we are creating a KO-model to characterize XLOC_091701 functionally to explore the regulatory mechanisms behind the observed favorable prognosis.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): AML, Molecular markers, Outcome


