
Contributions
Abstract: PB1595
Type: Publication Only
Background
Intragenic interstitial deletion of IKZF1 is a recurrent genomic alteration in B-cell acute lymphoblastic leukemia (B-ALL). The deletions are mediated by illegitimate V(D)J recombination via cryptic recombination signal sequences (RSSs). There were 4 major types of IKZF1 deletion (∆4-7, ∆2-7, ∆4-8, ∆2-8). The presence of IKZF1 mutation has been reported to be an independent risk factor of poor prognosis for patients with B-ALL. Despite the prognostic value, there are no suitable testing methods for detecting the mutation, and current clinical applications are therefore limited.
Aims
To detect various type of IKZF1 deletion including the commonly deleted exon 4-7 region, we developed a fluorescence in-situ hybridization (FISH) probe set. We validated the probes using clinical samples.
Methods
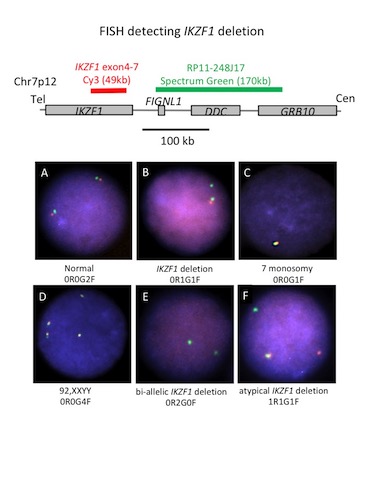
The probe set consists of a designed probe for the commonly deleted region (Cy3, red) and a bacterial artificial chromosomes (BAC) clone probe for detecting the 3’ flanking region (Spectrum Green; RP11-248J17). The IKZF1 commonly deleted region was amplified using long PCR method. Normal cutoff was validated using 10 peripheral blood samples from normal individuals. Twenty-three clinical samples (9 Ph(+)ALL, 8 Ph(-)ALL, 3 CML lymphoid crisis, 3 AML) were analyzed using the probe. IKZF1 deletion was also validated by multiplex PCR method and each deletion joint was sequence verified.
Results
Normal cell showed 2 fusion signal reflecting intact IKZF1 alleles (0R0G2F). In the cell with heterozygous IKZF1 deletion, intact IKZF1 allele showed a fusion signal, and the deleted allele showed loss of the red signal (0R1G1F). Normal cutoff value for IKZF1 deletion was established to be 1.5% from 10 normal samples. Gain or loss of entire IKZF1 region was detected by gain or loss of fusion signal such as 0R0G4F or 0R0G1F. Intragenic IKZF1 deletion was frequent in Ph(+)ALL (7/9, 77.8%) but relatively rare in Ph(-)ALL (3/8, 37.5%) and CML lymphoid crisis (1/3, 33.3%). On the other hand, IKZF1 deletion was not detected in any of the AML samples. For the cases with IKZF1 deletion, the exact deletion type could be identified by multiplex PCR in all cases. The joint sequence was unique to each patient showing a typical RAG1/2-mediated V(D)J recombination signature. One case showed 2 IKZF1 deletion signals in each cell (0R2G0F). Multiplex PCR verified 2 independent deletion (∆,2-7 ∆4-8) in the case.
One case showed an atypical break-apart signal (1R1G1F). Inverse PCR of the case revealed insertion of the deleted IKZF1 fragment into a legitimate RSSs site at immunoglobulin kappa on chromosome 2. This is the first description showing that the insertion of a RAG1/2-mediated fragment can occur between genuine and cryptic RSSs in ALL. RAG1/2-mediated genome exchange could play a role in leukemogenesis by causing cancer-associated insertion that results in disruption of the expression of a tumor suppressor.
Conclusion
In this study, we established FISH probes detecting IKZF1 deletion in a quick, quantitative and cost-effective manner. FISH based assay provided a novel insight into B-cell receptor editing by insertion of a cryptic RSS-mediated genomic fragment in ALL biology.
Session topic: 1. Acute lymphoblastic leukemia – Biology & Translational Research
Keyword(s): Acute lymphoblastic leukemia, FISH, Ikaros
Abstract: PB1595
Type: Publication Only
Background
Intragenic interstitial deletion of IKZF1 is a recurrent genomic alteration in B-cell acute lymphoblastic leukemia (B-ALL). The deletions are mediated by illegitimate V(D)J recombination via cryptic recombination signal sequences (RSSs). There were 4 major types of IKZF1 deletion (∆4-7, ∆2-7, ∆4-8, ∆2-8). The presence of IKZF1 mutation has been reported to be an independent risk factor of poor prognosis for patients with B-ALL. Despite the prognostic value, there are no suitable testing methods for detecting the mutation, and current clinical applications are therefore limited.
Aims
To detect various type of IKZF1 deletion including the commonly deleted exon 4-7 region, we developed a fluorescence in-situ hybridization (FISH) probe set. We validated the probes using clinical samples.
Methods
The probe set consists of a designed probe for the commonly deleted region (Cy3, red) and a bacterial artificial chromosomes (BAC) clone probe for detecting the 3’ flanking region (Spectrum Green; RP11-248J17). The IKZF1 commonly deleted region was amplified using long PCR method. Normal cutoff was validated using 10 peripheral blood samples from normal individuals. Twenty-three clinical samples (9 Ph(+)ALL, 8 Ph(-)ALL, 3 CML lymphoid crisis, 3 AML) were analyzed using the probe. IKZF1 deletion was also validated by multiplex PCR method and each deletion joint was sequence verified.
Results
Normal cell showed 2 fusion signal reflecting intact IKZF1 alleles (0R0G2F). In the cell with heterozygous IKZF1 deletion, intact IKZF1 allele showed a fusion signal, and the deleted allele showed loss of the red signal (0R1G1F). Normal cutoff value for IKZF1 deletion was established to be 1.5% from 10 normal samples. Gain or loss of entire IKZF1 region was detected by gain or loss of fusion signal such as 0R0G4F or 0R0G1F. Intragenic IKZF1 deletion was frequent in Ph(+)ALL (7/9, 77.8%) but relatively rare in Ph(-)ALL (3/8, 37.5%) and CML lymphoid crisis (1/3, 33.3%). On the other hand, IKZF1 deletion was not detected in any of the AML samples. For the cases with IKZF1 deletion, the exact deletion type could be identified by multiplex PCR in all cases. The joint sequence was unique to each patient showing a typical RAG1/2-mediated V(D)J recombination signature. One case showed 2 IKZF1 deletion signals in each cell (0R2G0F). Multiplex PCR verified 2 independent deletion (∆,2-7 ∆4-8) in the case.
One case showed an atypical break-apart signal (1R1G1F). Inverse PCR of the case revealed insertion of the deleted IKZF1 fragment into a legitimate RSSs site at immunoglobulin kappa on chromosome 2. This is the first description showing that the insertion of a RAG1/2-mediated fragment can occur between genuine and cryptic RSSs in ALL. RAG1/2-mediated genome exchange could play a role in leukemogenesis by causing cancer-associated insertion that results in disruption of the expression of a tumor suppressor.

Conclusion
In this study, we established FISH probes detecting IKZF1 deletion in a quick, quantitative and cost-effective manner. FISH based assay provided a novel insight into B-cell receptor editing by insertion of a cryptic RSS-mediated genomic fragment in ALL biology.
Session topic: 1. Acute lymphoblastic leukemia – Biology & Translational Research
Keyword(s): Acute lymphoblastic leukemia, FISH, Ikaros


