
Contributions
Abstract: PB1988
Type: Publication Only
Background
The RUNX1 gene encodes a transcription factor and is a key regulator in the development of normal hematopoiesis. Somatic RUNX1 mutations are recurrent in malignant diseases including myelodysplastic syndromes (MDS), acute myeloid leukemia (AML) and germline RUNX1 mutations are associated with Familial platelet disorder with predisposition to AML (FPD/AML). In this study we analyzed serial samples from a 10-year-old girl with prolonged thrombocytopenia post AML treatment for a constitutional RUNX1 mutation. A remission PB sample 510 days post AML diagnosis was investigated by targeted NGS and showed a RUNX1 c.347T>C (p. F116S) mutation which was detected at a variant allele frequency (VAF) of 6%.
Aims
Establishment of a ddPCR assay for detection of a mosaic RUNX1 mutation and exploration of its longitudinal evolution and presence in different hematopoietic cell lineages.
Methods
A droplet digital PCR (ddPCR) assay was designed for detection of the RUNX1 c.347C>T mutation. Primers and HEX/FAM labeled probes were designed for detection of both mutant and wild type DNA. The assay sensitivity was experimentally validated to be 0.1%. For sequential studies DNA was isolated from peripheral blood (PB) and bone marrow (BM) remission samples. T-cells, B-cells, and myeloid cells were isolated from PB with CD3, CD19, and CD15 antibodies respectively using EasySepTM reagents from Stem Cell Technology. Fibroblasts and keratinocytes from a skin biopsy were cultivated for further analysis, and DNA from the patient’s Guthrie card was extracted.
Results
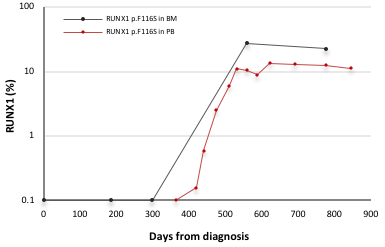
Fibroblasts and keratinocytes from the skin biopsy were found to be negative for the RUNX1 mutation. While positive for GATA2 and double CEBPA mutations at AML diagnosis no RUNX1 mutation could be detected in the diagnostic sample. In sequential analyses of PB and BM post AML the mutation was seen for the first time in PB 419 days post AML with a VAF of 0.2%, increasing to 13% and 23%, over the next 400 days (figure). Analyses of T-cells, B-cells, and myeloid cells isolated from a PB remission sample 845 days post AML diagnosis showed presence of the RUNX1 mutation in all three lineages with allele frequencies of 4%, 32%, and 8% respectively.
Conclusion
The RUNX1 mutation identified was excluded as being germline due to its absence in fibroblast and keratinocytes from skin. Using ddPCR we were able to detect the mutation in T-cells, B-cells, and myeloid cells meaning that the mutation has originated in a hematopoietic stem cell. The patient was reported to have had a lifelong tendency to easy bruising and it is tempting to speculate that the mutation occurred in the hematopoietic system during embryonic development. However, we were not able to detect the mutation in DNA from patient's Guthrie card and have no other PB samples prior to the AML diagnosis. If the RUNX1 mutation is responsible for the reported easy bruising and post AML thrombocytopenia, it must arise from another hematopoietic stem cell than those leading to the AML. Another possibility is that the RUNX1 mutation is therapy induced and possesses a proliferative advantage compared to normal stem cells.
Future fluorescence activated cell sorting of normal and leukemic hematopoietic stem cells from the diagnostic AML sample and subsequent RUNX1 ddPCR analysis will hopefully aid us in determining the occurrence of the mutation.
Session topic: 24. Hematopoiesis, stem cells and microenvironment
Keyword(s): mutation analysis, RUNX1, Stem cell
Abstract: PB1988
Type: Publication Only
Background
The RUNX1 gene encodes a transcription factor and is a key regulator in the development of normal hematopoiesis. Somatic RUNX1 mutations are recurrent in malignant diseases including myelodysplastic syndromes (MDS), acute myeloid leukemia (AML) and germline RUNX1 mutations are associated with Familial platelet disorder with predisposition to AML (FPD/AML). In this study we analyzed serial samples from a 10-year-old girl with prolonged thrombocytopenia post AML treatment for a constitutional RUNX1 mutation. A remission PB sample 510 days post AML diagnosis was investigated by targeted NGS and showed a RUNX1 c.347T>C (p. F116S) mutation which was detected at a variant allele frequency (VAF) of 6%.
Aims
Establishment of a ddPCR assay for detection of a mosaic RUNX1 mutation and exploration of its longitudinal evolution and presence in different hematopoietic cell lineages.
Methods
A droplet digital PCR (ddPCR) assay was designed for detection of the RUNX1 c.347C>T mutation. Primers and HEX/FAM labeled probes were designed for detection of both mutant and wild type DNA. The assay sensitivity was experimentally validated to be 0.1%. For sequential studies DNA was isolated from peripheral blood (PB) and bone marrow (BM) remission samples. T-cells, B-cells, and myeloid cells were isolated from PB with CD3, CD19, and CD15 antibodies respectively using EasySepTM reagents from Stem Cell Technology. Fibroblasts and keratinocytes from a skin biopsy were cultivated for further analysis, and DNA from the patient’s Guthrie card was extracted.
Results
Fibroblasts and keratinocytes from the skin biopsy were found to be negative for the RUNX1 mutation. While positive for GATA2 and double CEBPA mutations at AML diagnosis no RUNX1 mutation could be detected in the diagnostic sample. In sequential analyses of PB and BM post AML the mutation was seen for the first time in PB 419 days post AML with a VAF of 0.2%, increasing to 13% and 23%, over the next 400 days (figure). Analyses of T-cells, B-cells, and myeloid cells isolated from a PB remission sample 845 days post AML diagnosis showed presence of the RUNX1 mutation in all three lineages with allele frequencies of 4%, 32%, and 8% respectively.

Conclusion
The RUNX1 mutation identified was excluded as being germline due to its absence in fibroblast and keratinocytes from skin. Using ddPCR we were able to detect the mutation in T-cells, B-cells, and myeloid cells meaning that the mutation has originated in a hematopoietic stem cell. The patient was reported to have had a lifelong tendency to easy bruising and it is tempting to speculate that the mutation occurred in the hematopoietic system during embryonic development. However, we were not able to detect the mutation in DNA from patient's Guthrie card and have no other PB samples prior to the AML diagnosis. If the RUNX1 mutation is responsible for the reported easy bruising and post AML thrombocytopenia, it must arise from another hematopoietic stem cell than those leading to the AML. Another possibility is that the RUNX1 mutation is therapy induced and possesses a proliferative advantage compared to normal stem cells.
Future fluorescence activated cell sorting of normal and leukemic hematopoietic stem cells from the diagnostic AML sample and subsequent RUNX1 ddPCR analysis will hopefully aid us in determining the occurrence of the mutation.
Session topic: 24. Hematopoiesis, stem cells and microenvironment
Keyword(s): mutation analysis, RUNX1, Stem cell


