
Contributions
Abstract: PB1662
Type: Publication Only
Background
With the advent of NGS, an extended spectrum of molecular abnormalities has been discovered. However, the correlations between most somatic mutations and clinical characteristics and the underlying mechanisms still need further investigations. In AML, several mutations (FLT3-ITD, NMP1, DNMT3, CEBPA, TET2, IDH1/2) are already described with known implications in prognosis and clinical evolution. However, little is known about CALR and CBL mutations in AML and MDS. As known, Calreticulin (CALR) plays important roles in calcium homeostasis, protein folding, cell proliferation, apoptosis and inmune response. CALR is the second most frequently mutated gene in myeloproliferative neoplasms (MPNs). Casitas B-cell lymphoma (CBL) is an E3 ubiquitin ligase and promotes ubiquitination-directed degradation of target proteins (EGFR, FLT3, KIT, MPL and Src). Loss of E3 ligase activity together with additional gain-of-functions induced by these mutations promote malignant transformation.
Aims
To report and to describe the frequency of atypical mutations in AML ans MDS, focused on CBL and CALR detected by NGS myeloid panel.
Methods
We performed a custom-targeted sequencing panel of 32 genes (all coding regions) implicated in leukemia prognosis, including ASXL1, CBL, DNMT3A, EPOR, ETV6, EZH2, FLT3, HRAS, IDH1, IDH2, JAK2, KDM6A, KIT, KRAS, LNK, MLL, MPL, NRAS, PHF6, PRPF40B, PTEN, RUNX1, SF1, SF3A1, SF3B1, SRSF2, TET2, TP53, U2AF35, VHL, ZRSR2 and CALR, by Ion Torrent Proton System-Thermo Fisher. Kaplan- Meier survival curves and Long-rank test were used for estimation of survival and difference between groups.DNA samples collected between 2012 to 2018 years from a cohort of 335 patients with myeloid pathologies were screened by NGS. Among all studied patients, 190 (53,5%) were male and 165 (46,5%) female. 231 (65,1%) had AML diagnosis, 77 (21,7%) myelodisplasic syndrome (MDS), 42 (11,8%) chronic myeloproliferative syndrome (NMP), 3 (0,8%) medular aplasia and 2 (0,6%) other pathologies.
Results
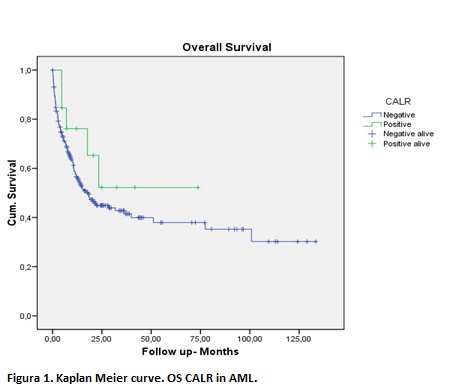
Out of 77 patients with MDS, 8 (10%) were positive for CBL and 20 (26%) for CALR mutation. There were 20 deaths in the MDS group, none of them had CALR mutation and just one patient had CBL mutation. In NMPS, 2 (4%) patients were positive for CBL and 4 (10%) for CALR mutation. In AML cohort, 13 (5,6%) were CALR positive and 15 (6,5%) CBL positive. 114 (49,5%) of AML patients were deceased, of which 5 (4%) were CALR and 9 (8%) CBL positives These results were not statistically significant (p >0.05). We observed an unusual location of CALR mutation, mostly SNV in exon 5, instead of indels in exon 9, the typical location in NMPs. We detected a hotspot location of mutations in the ring finger domain of CBL similar to the ones described in previous publications. Median OS were 18 months for CALR negative AML group vs not reached median OS for CALR positive AML group (p=NS). Median OS were 71 months for CALR negative MDS group vs 108 months for CALR positive MDS group (p=NS)
Conclusion
These results reveal a possible correlation between CALR gene mutation and prognosis features in AML or MDS patients. Despite of the lack of statistical significance, routine diagnostic approach of this gene could improve prognostic prediction. Moreover, it could be a therapeutic target, leading us to an individualized treatment. On the other hand, we could not find any correlation betweeen CBL mutation and MDS or AML and its prognosis. Therefore, larger controlled studies are necessary to confirm the findings.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): Acute Myeloid Leukemia, Molecular markers, mutation analysis, Myelodysplasia
Abstract: PB1662
Type: Publication Only
Background
With the advent of NGS, an extended spectrum of molecular abnormalities has been discovered. However, the correlations between most somatic mutations and clinical characteristics and the underlying mechanisms still need further investigations. In AML, several mutations (FLT3-ITD, NMP1, DNMT3, CEBPA, TET2, IDH1/2) are already described with known implications in prognosis and clinical evolution. However, little is known about CALR and CBL mutations in AML and MDS. As known, Calreticulin (CALR) plays important roles in calcium homeostasis, protein folding, cell proliferation, apoptosis and inmune response. CALR is the second most frequently mutated gene in myeloproliferative neoplasms (MPNs). Casitas B-cell lymphoma (CBL) is an E3 ubiquitin ligase and promotes ubiquitination-directed degradation of target proteins (EGFR, FLT3, KIT, MPL and Src). Loss of E3 ligase activity together with additional gain-of-functions induced by these mutations promote malignant transformation.
Aims
To report and to describe the frequency of atypical mutations in AML ans MDS, focused on CBL and CALR detected by NGS myeloid panel.
Methods
We performed a custom-targeted sequencing panel of 32 genes (all coding regions) implicated in leukemia prognosis, including ASXL1, CBL, DNMT3A, EPOR, ETV6, EZH2, FLT3, HRAS, IDH1, IDH2, JAK2, KDM6A, KIT, KRAS, LNK, MLL, MPL, NRAS, PHF6, PRPF40B, PTEN, RUNX1, SF1, SF3A1, SF3B1, SRSF2, TET2, TP53, U2AF35, VHL, ZRSR2 and CALR, by Ion Torrent Proton System-Thermo Fisher. Kaplan- Meier survival curves and Long-rank test were used for estimation of survival and difference between groups.DNA samples collected between 2012 to 2018 years from a cohort of 335 patients with myeloid pathologies were screened by NGS. Among all studied patients, 190 (53,5%) were male and 165 (46,5%) female. 231 (65,1%) had AML diagnosis, 77 (21,7%) myelodisplasic syndrome (MDS), 42 (11,8%) chronic myeloproliferative syndrome (NMP), 3 (0,8%) medular aplasia and 2 (0,6%) other pathologies.
Results
Out of 77 patients with MDS, 8 (10%) were positive for CBL and 20 (26%) for CALR mutation. There were 20 deaths in the MDS group, none of them had CALR mutation and just one patient had CBL mutation. In NMPS, 2 (4%) patients were positive for CBL and 4 (10%) for CALR mutation. In AML cohort, 13 (5,6%) were CALR positive and 15 (6,5%) CBL positive. 114 (49,5%) of AML patients were deceased, of which 5 (4%) were CALR and 9 (8%) CBL positives These results were not statistically significant (p >0.05). We observed an unusual location of CALR mutation, mostly SNV in exon 5, instead of indels in exon 9, the typical location in NMPs. We detected a hotspot location of mutations in the ring finger domain of CBL similar to the ones described in previous publications. Median OS were 18 months for CALR negative AML group vs not reached median OS for CALR positive AML group (p=NS). Median OS were 71 months for CALR negative MDS group vs 108 months for CALR positive MDS group (p=NS)

Conclusion
These results reveal a possible correlation between CALR gene mutation and prognosis features in AML or MDS patients. Despite of the lack of statistical significance, routine diagnostic approach of this gene could improve prognostic prediction. Moreover, it could be a therapeutic target, leading us to an individualized treatment. On the other hand, we could not find any correlation betweeen CBL mutation and MDS or AML and its prognosis. Therefore, larger controlled studies are necessary to confirm the findings.
Session topic: 3. Acute myeloid leukemia - Biology & Translational Research
Keyword(s): Acute Myeloid Leukemia, Molecular markers, mutation analysis, Myelodysplasia


