
Contributions
Abstract: S1571
Type: Oral Presentation
Presentation during EHA23: On Sunday, June 17, 2018 from 08:15 - 08:30
Location: Room A6
Background
Even in the era of the targeted therapies, there remains clinical value in exploring the impact of disease characteristics in chemo-immunotherapy (CIT) trials, namely to identify features that contribute most to long-term outcomes, thereby pinpointing patients destined to benefit from these therapies. One such feature is the CLL DNA methylome, that recapitulates normal B cell maturation, with IGHV mutated (M-CLL) and unmutated CLL (U-CLL) retaining an imprint of the DNA methylation signature of memory (m-CLL) and naive B cells (n-CLL), respectively, with a third intermediate epigenetic subgroup (i-CLL) with borderline mutation status. The pyrosequencing analysis of 5 CpG sites can divide CLL into these three subgroups, each with different clinico-biological features.
Aims
To validate the clinical utility of the epigenetic subgroups in patients entering clinical trials.
Methods
We classified 605 treatment-naïve patients randomized to one chemotherapy (CLL4) and two CIT (ARCTIC and ADMIRE) trials, into the three epigenetic subgroups using pyrosequencing and confirmatory Infinium HumanMethylation450 analysis (n=60, 96% concordance between technologies).
Results
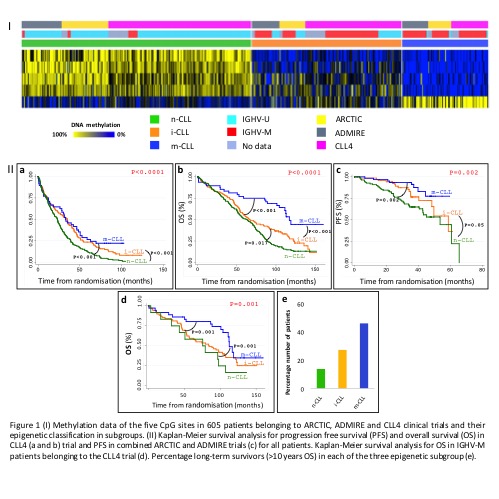
We identified n-, i- and m-CLL in 49.5% (n=298), 32.0% (n=195) and 18.5% (n=112) of our patients, respectively (Fig1I). Fewer m-CLL patients were identified in our study compared to published data, reflecting the progressive nature of our cohort, with 80% (n=245/305, P<0.001) of U-CLL cases exhibited the n-CLL signature (i-CLL: 17% and m-CLL: 3%). For M-CLL cases 9%, 50% and 41% exhibited the n-, i- and m-CLL epigenetic signature, respectively. In 359 CLL4 patients, n-, i- and m-CLL patients exhibited a median PFS of 23, 35 and 33, and OS of 63, 66 and 106 months, respectively (Fig1IIa and b). n-CLL showed significantly shorter PFS than i-CLL (HR 0.64, p<0.001) and m-CLL (HR 0.52, p<0.001), and had the shortest OS, again compared to i-CLL (HR 0.73, p=0.01) and m-CLL (HR 0.33, p<0.001). The epigenetic signature was associated with long-term survival (>10 years OS), with n-CLL accounting for only 14% of this subgroup (P<0.001, FigIIe). In IGHV-M patients, the m-CLL subgroup showed 104 months of median OS compared to 79 and 84 months in n- and i-CLL subgroups (FigIId). Multivariate Cox proportional analysis, controlling for confounding variables (inc. clinical features, IGHV status, TP53, NOTCH1 and SF3B1) in 278 patients showed that m-CLL was an independent prognostic factor for OS (HR 0.46, p<0.01), but not PFS. In univariate analysis, in 247 patients randomized to ARCTIC and ADMIRE, the i- (HR:0.57, p=0.05) and m-CLL (HR:0.3, p=0.002) subgroups displayed longer PFS (Fig1IIc). In a multivariate model, including TP53 lesions and IGHV status (239 patients), the m-CLL subgroup retained its independent prognostic significance (HR:0.25, p<0.001).
Conclusion
In conclusion, we report the first analysis of the clinical utility of epigenetic subgroup signatures in patients entered into first-line chemotherapy and CIT trials and identified m-CLL as an independent marker of survival in both our CLL4 and ARCTIC/ADMIRE cohorts. This provides important evidence that DNA methylation analysis may aid in the identification of patients destined to demonstrate durable remissions when treated with these agents.
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, DNA methylation, prognosis
Abstract: S1571
Type: Oral Presentation
Presentation during EHA23: On Sunday, June 17, 2018 from 08:15 - 08:30
Location: Room A6
Background
Even in the era of the targeted therapies, there remains clinical value in exploring the impact of disease characteristics in chemo-immunotherapy (CIT) trials, namely to identify features that contribute most to long-term outcomes, thereby pinpointing patients destined to benefit from these therapies. One such feature is the CLL DNA methylome, that recapitulates normal B cell maturation, with IGHV mutated (M-CLL) and unmutated CLL (U-CLL) retaining an imprint of the DNA methylation signature of memory (m-CLL) and naive B cells (n-CLL), respectively, with a third intermediate epigenetic subgroup (i-CLL) with borderline mutation status. The pyrosequencing analysis of 5 CpG sites can divide CLL into these three subgroups, each with different clinico-biological features.
Aims
To validate the clinical utility of the epigenetic subgroups in patients entering clinical trials.
Methods
We classified 605 treatment-naïve patients randomized to one chemotherapy (CLL4) and two CIT (ARCTIC and ADMIRE) trials, into the three epigenetic subgroups using pyrosequencing and confirmatory Infinium HumanMethylation450 analysis (n=60, 96% concordance between technologies).
Results
We identified n-, i- and m-CLL in 49.5% (n=298), 32.0% (n=195) and 18.5% (n=112) of our patients, respectively (Fig1I). Fewer m-CLL patients were identified in our study compared to published data, reflecting the progressive nature of our cohort, with 80% (n=245/305, P<0.001) of U-CLL cases exhibited the n-CLL signature (i-CLL: 17% and m-CLL: 3%). For M-CLL cases 9%, 50% and 41% exhibited the n-, i- and m-CLL epigenetic signature, respectively. In 359 CLL4 patients, n-, i- and m-CLL patients exhibited a median PFS of 23, 35 and 33, and OS of 63, 66 and 106 months, respectively (Fig1IIa and b). n-CLL showed significantly shorter PFS than i-CLL (HR 0.64, p<0.001) and m-CLL (HR 0.52, p<0.001), and had the shortest OS, again compared to i-CLL (HR 0.73, p=0.01) and m-CLL (HR 0.33, p<0.001). The epigenetic signature was associated with long-term survival (>10 years OS), with n-CLL accounting for only 14% of this subgroup (P<0.001, FigIIe). In IGHV-M patients, the m-CLL subgroup showed 104 months of median OS compared to 79 and 84 months in n- and i-CLL subgroups (FigIId). Multivariate Cox proportional analysis, controlling for confounding variables (inc. clinical features, IGHV status, TP53, NOTCH1 and SF3B1) in 278 patients showed that m-CLL was an independent prognostic factor for OS (HR 0.46, p<0.01), but not PFS. In univariate analysis, in 247 patients randomized to ARCTIC and ADMIRE, the i- (HR:0.57, p=0.05) and m-CLL (HR:0.3, p=0.002) subgroups displayed longer PFS (Fig1IIc). In a multivariate model, including TP53 lesions and IGHV status (239 patients), the m-CLL subgroup retained its independent prognostic significance (HR:0.25, p<0.001).

Conclusion
In conclusion, we report the first analysis of the clinical utility of epigenetic subgroup signatures in patients entered into first-line chemotherapy and CIT trials and identified m-CLL as an independent marker of survival in both our CLL4 and ARCTIC/ADMIRE cohorts. This provides important evidence that DNA methylation analysis may aid in the identification of patients destined to demonstrate durable remissions when treated with these agents.
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, DNA methylation, prognosis


