
Contributions
Abstract: S879
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:45 - 17:00
Location: Room A7
Background
Myelodysplastic syndromes (MDS) are uncommon in children. There are limited known on the gene mutations landscape in pediatric patients with MDS and the differences between pediatric and adult patients.
Aims
We aimed to elucidate the mutational landscape in pediatric MDS and compared with adult cases.
Methods
We performed both whole exome sequencing and targeted sequencing on 30 pediatric MDS patients and targeted sequencing on 57 pediatric cases and 510 adult patients. The diagnosis was according to the 2016 revised criteria of the WHO for childhood MDS. To be comparable with adult and on the basis of difficulty of precisely diagnosing of low-risk RCC, only pediatric patients who also meet the diagnostic criteria for adult MDS were included. DNA was obtained from bone marrow. Germline status was analyzed by CD3+ T-cell or oral epithelial cells.
Results
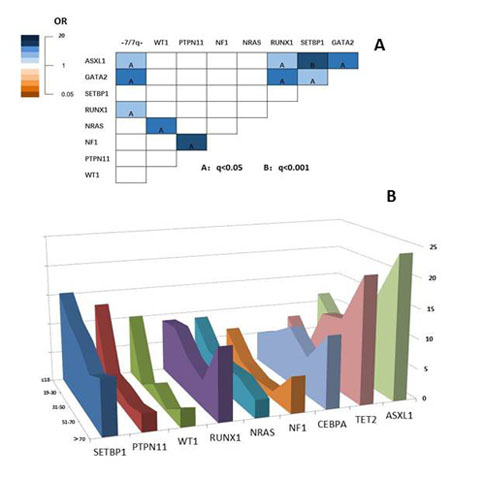
We found 208 mutations in 54 distinct genes with variant allelic frequency (VAF) from 2% to 100%. 83% patients had at least 1 gene mutation. 11 genes were mutated in more than 5% of patients, including SETBP1 15%, PTPN11 13%, ASXL1 10%, WT1 9%, RUNX1 8%, NRAS 8%, TET2 7%, NF1 7%, CEBPA 5%, GATA1 5%, FLT3 5%. Germline mutation of GATA2 and SAMD9/SAMD9L were detected in 9% and 5% of the patients. Mutation of DNMT3A, TP53, SF3B1, SRSF2 were rare (1%, 2%, 0%, 2%,). The most frequent pathways were the epigenetic modification (37%) and the RAS pathway (30%). In univariate analysis, somatic mutated SETBP1, ASXL1, RUNX1 and deletion of chromosome 7 were significantly enriched in germline GATA2 mutated subjects (P<0.05). And NRAS and NF1 were significantly associated with WT1 and PTPN11 mutated subjects, respectively (Figure 1A). The multivariate analysis showed ASXL1 was significantly enriched in GATA2 mutated subjects, and NRAS and NF1 were significantly associated with WT1 and PTPN11 subjects, respectively (P<0.05). According to the above results, the cohort could be divided into three subgroups, namely GATA2/ASXL1/SETBP1/RUNX1 group, the WT1/NRAS group and the PTPN11/NF1 group. Using copy number-adjusted VAF, we reconstructed the clonal architecture to explore ancestral and subclonal mutations. ASXL1, NRAS, and PTPN11 were more likely to be an ancestral mutation. The 5-year overall survival (OS) was 57%. For patients with and without HSCT, the 5-year OS was 92% and 40% (P<0.001). Patients with deletion of chromosome 7, mutation of PTPN11 and SETBP1 tend to have worse OS. However, for patients accepted HSCT, there was no significant association between the genetic mutation and OS. In adult patients from our institution, the most common mutated genes were spliceosome and epigenetic associated genes. Pediatric patients had higher VAF than adult patients in SETBP1, PTPN11, RUNX1, TET2, CEBPA ( the average VAF was 37.4% v.s. 20.2%; 38.3% v.s.18.6%; 38.3% v.s.18.6%; 44.0% v.s. 22.9%; 43.7% v.s. 17.0%; 36.2% v.s. 16.2%, P<0.001).We also found that SETBP1, PTPN11, WT1, FLT3 more frequently occurred in pediatric and young adult patients, while, TET2 and ASXL1 were more likely existing in patients more than 70 years old (Figure 1B).
Conclusion
In this study, we found that mutations frequent encountered in adult MDS and in age-related clonal hematopoiesis were rare in children. We also verified that germline mutations like GATA2 and SAMD9/SAMD9L are common in pediatric patients. Somatic mutations in SETBP1, ASXL1, RUNX1, WT1 and the RAS oncogenes define the genomic landscape of the pediatric MDS. Collectively, our results help define the mutational landscape in MDS in children.
Session topic: 10. Myelodysplastic syndromes – Clinical
Keyword(s): mutation analysis, Myelodysplasia, Pediatric
Abstract: S879
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:45 - 17:00
Location: Room A7
Background
Myelodysplastic syndromes (MDS) are uncommon in children. There are limited known on the gene mutations landscape in pediatric patients with MDS and the differences between pediatric and adult patients.
Aims
We aimed to elucidate the mutational landscape in pediatric MDS and compared with adult cases.
Methods
We performed both whole exome sequencing and targeted sequencing on 30 pediatric MDS patients and targeted sequencing on 57 pediatric cases and 510 adult patients. The diagnosis was according to the 2016 revised criteria of the WHO for childhood MDS. To be comparable with adult and on the basis of difficulty of precisely diagnosing of low-risk RCC, only pediatric patients who also meet the diagnostic criteria for adult MDS were included. DNA was obtained from bone marrow. Germline status was analyzed by CD3+ T-cell or oral epithelial cells.
Results
We found 208 mutations in 54 distinct genes with variant allelic frequency (VAF) from 2% to 100%. 83% patients had at least 1 gene mutation. 11 genes were mutated in more than 5% of patients, including SETBP1 15%, PTPN11 13%, ASXL1 10%, WT1 9%, RUNX1 8%, NRAS 8%, TET2 7%, NF1 7%, CEBPA 5%, GATA1 5%, FLT3 5%. Germline mutation of GATA2 and SAMD9/SAMD9L were detected in 9% and 5% of the patients. Mutation of DNMT3A, TP53, SF3B1, SRSF2 were rare (1%, 2%, 0%, 2%,). The most frequent pathways were the epigenetic modification (37%) and the RAS pathway (30%). In univariate analysis, somatic mutated SETBP1, ASXL1, RUNX1 and deletion of chromosome 7 were significantly enriched in germline GATA2 mutated subjects (P<0.05). And NRAS and NF1 were significantly associated with WT1 and PTPN11 mutated subjects, respectively (Figure 1A). The multivariate analysis showed ASXL1 was significantly enriched in GATA2 mutated subjects, and NRAS and NF1 were significantly associated with WT1 and PTPN11 subjects, respectively (P<0.05). According to the above results, the cohort could be divided into three subgroups, namely GATA2/ASXL1/SETBP1/RUNX1 group, the WT1/NRAS group and the PTPN11/NF1 group. Using copy number-adjusted VAF, we reconstructed the clonal architecture to explore ancestral and subclonal mutations. ASXL1, NRAS, and PTPN11 were more likely to be an ancestral mutation. The 5-year overall survival (OS) was 57%. For patients with and without HSCT, the 5-year OS was 92% and 40% (P<0.001). Patients with deletion of chromosome 7, mutation of PTPN11 and SETBP1 tend to have worse OS. However, for patients accepted HSCT, there was no significant association between the genetic mutation and OS. In adult patients from our institution, the most common mutated genes were spliceosome and epigenetic associated genes. Pediatric patients had higher VAF than adult patients in SETBP1, PTPN11, RUNX1, TET2, CEBPA ( the average VAF was 37.4% v.s. 20.2%; 38.3% v.s.18.6%; 38.3% v.s.18.6%; 44.0% v.s. 22.9%; 43.7% v.s. 17.0%; 36.2% v.s. 16.2%, P<0.001).We also found that SETBP1, PTPN11, WT1, FLT3 more frequently occurred in pediatric and young adult patients, while, TET2 and ASXL1 were more likely existing in patients more than 70 years old (Figure 1B).

Conclusion
In this study, we found that mutations frequent encountered in adult MDS and in age-related clonal hematopoiesis were rare in children. We also verified that germline mutations like GATA2 and SAMD9/SAMD9L are common in pediatric patients. Somatic mutations in SETBP1, ASXL1, RUNX1, WT1 and the RAS oncogenes define the genomic landscape of the pediatric MDS. Collectively, our results help define the mutational landscape in MDS in children.
Session topic: 10. Myelodysplastic syndromes – Clinical
Keyword(s): mutation analysis, Myelodysplasia, Pediatric


