
Contributions
Abstract: S869
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:45 - 17:00
Location: Room K1
Background
The B-cell receptor (BCR) is one of the most important surface molecules that CLL cells use to gain oncogenic signals from the microenvironment. The critical role of BCR signaling for the pathogenesis of CLL is supported by the therapeutic success of ibrutinib, a targeted agent that disrupts the BCR pathway. Beside microenvironment-promoted oncogenic signals, the biology of CLL is also driven by molecular lesions and clonal evolution, that mark CLL progression and treatment resistance. The interconnection between microenvironment-promoted oncogenic signals and clonal evolution has been postulated in CLL but never proven because of the lack of suitable ex vivo models.
Aims
Ibrutinib allows the unprecedented opportunity of assessing the contribution of BCR to cancer clonal evolution directly in vivo in patients.
Methods
The IOSI-EMA-001 study (NCT02827617) is an observational, non-interventional, multicenter study consisting in the prospective and longitudinal collection of peripheral blood samples and clinical data from high risk CLL patients treated with ibrutinib monotherapy. Tumor DNA derived from sorted CLL cells (purity >99%) and germline DNA derived from sorted T cells were used for somatic mutation identification by CAPP-seq targeted deep next generation. A gene panel including 133 genes recurrently mutated in mature B-cell tumors or targeted by the aberrant somatic hypermutation process, was used. The number of the libraries loaded in the NexSeq500 sequencer (Illumina) was tailored at obtaining at least a coverage >2000x in >80% of the region of interest. A stringent bioinformatic pipeline was applied to suppress the background noise allowing to call variants with a sensitivity of 3x10-3. To track clonal trajectories across serial samples, we first measured the variant allele fraction (VAF) of all mutations identified across the different timepoints per patient. VAFs were transformed to cancer cell fractions (CCFs) using the ABSOLUTE tool. For each patient, we compared the clonal composition of the baseline sample with all the available longitudinal samples (up to 72 weeks of therapy).
Results
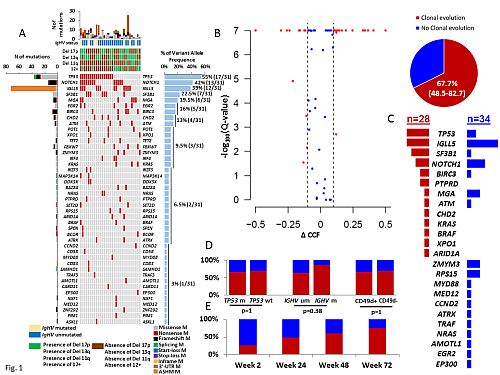
The study cohort comprised 31 high risk CLL patients, including 15 treatment naïve, 16 relapsed, 80% IGHV unmutated, 42% 17p deleted and 55% TP53 mutated (Fig. 1A). Median duration of ibrutinib treatment was 45 weeks (range 24-72 weeks). Overall, 285 individual mutations were longitudinally discovered and monitored across a total of 119 sequential timepoints collected during ibrutinib treatment. Significant changes in CCF over time, defined as a FDR adjusted p value of <0.1 for change in CCF >0.1 in the largest rising or falling clone was observed in 21/31 (67.7%) cases (Fig. 1B), a proportion that is superimposable to the clonal evolution rate previously documented in CLL treated with chemoimmunotherapy. Clonal evolution appeared to be puzzled and involved different genes without a stereotypic targeting (Fig. 1C). Consistently, none of the main driver gene mutations was homogeneously selected or suppressed by ibrutinib. Clonal evolution rate neither associated with IGHV or TP53 mutation status (Fig. 1D), nor changed over time (Fig. 1E).
Conclusion
Our results suggest that clonal evolution, a known pathogenic mechanism of progressive CLL: i) is not abrogated by ibrutinib; and ii) is quantitatively similar, but qualitatively different, than clonal evolution under chemoimmunotherapy, without specific pathways being targeted.
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, ibrutinib, mutation analysis
Abstract: S869
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:45 - 17:00
Location: Room K1
Background
The B-cell receptor (BCR) is one of the most important surface molecules that CLL cells use to gain oncogenic signals from the microenvironment. The critical role of BCR signaling for the pathogenesis of CLL is supported by the therapeutic success of ibrutinib, a targeted agent that disrupts the BCR pathway. Beside microenvironment-promoted oncogenic signals, the biology of CLL is also driven by molecular lesions and clonal evolution, that mark CLL progression and treatment resistance. The interconnection between microenvironment-promoted oncogenic signals and clonal evolution has been postulated in CLL but never proven because of the lack of suitable ex vivo models.
Aims
Ibrutinib allows the unprecedented opportunity of assessing the contribution of BCR to cancer clonal evolution directly in vivo in patients.
Methods
The IOSI-EMA-001 study (NCT02827617) is an observational, non-interventional, multicenter study consisting in the prospective and longitudinal collection of peripheral blood samples and clinical data from high risk CLL patients treated with ibrutinib monotherapy. Tumor DNA derived from sorted CLL cells (purity >99%) and germline DNA derived from sorted T cells were used for somatic mutation identification by CAPP-seq targeted deep next generation. A gene panel including 133 genes recurrently mutated in mature B-cell tumors or targeted by the aberrant somatic hypermutation process, was used. The number of the libraries loaded in the NexSeq500 sequencer (Illumina) was tailored at obtaining at least a coverage >2000x in >80% of the region of interest. A stringent bioinformatic pipeline was applied to suppress the background noise allowing to call variants with a sensitivity of 3x10-3. To track clonal trajectories across serial samples, we first measured the variant allele fraction (VAF) of all mutations identified across the different timepoints per patient. VAFs were transformed to cancer cell fractions (CCFs) using the ABSOLUTE tool. For each patient, we compared the clonal composition of the baseline sample with all the available longitudinal samples (up to 72 weeks of therapy).
Results
The study cohort comprised 31 high risk CLL patients, including 15 treatment naïve, 16 relapsed, 80% IGHV unmutated, 42% 17p deleted and 55% TP53 mutated (Fig. 1A). Median duration of ibrutinib treatment was 45 weeks (range 24-72 weeks). Overall, 285 individual mutations were longitudinally discovered and monitored across a total of 119 sequential timepoints collected during ibrutinib treatment. Significant changes in CCF over time, defined as a FDR adjusted p value of <0.1 for change in CCF >0.1 in the largest rising or falling clone was observed in 21/31 (67.7%) cases (Fig. 1B), a proportion that is superimposable to the clonal evolution rate previously documented in CLL treated with chemoimmunotherapy. Clonal evolution appeared to be puzzled and involved different genes without a stereotypic targeting (Fig. 1C). Consistently, none of the main driver gene mutations was homogeneously selected or suppressed by ibrutinib. Clonal evolution rate neither associated with IGHV or TP53 mutation status (Fig. 1D), nor changed over time (Fig. 1E).

Conclusion
Our results suggest that clonal evolution, a known pathogenic mechanism of progressive CLL: i) is not abrogated by ibrutinib; and ii) is quantitatively similar, but qualitatively different, than clonal evolution under chemoimmunotherapy, without specific pathways being targeted.
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, ibrutinib, mutation analysis


