
Contributions
Abstract: S826
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 12:15 - 12:30
Location: Room A6
Background
Chromosomal abnormalities are established prognostic markers in adult ALL. In the UKALL14 clinical trial patients with BCR-ABL1, KMT2A-AFF1, low hypodiploidy or a complex karyotype were classified as having high risk (HR) genetics. All remaining patients, including those lacking an established chromosomal abnormality (B-other ALL) were classified as having standard risk (HR) genetics. Recent genomic studies have identified a plethora of new gene fusions within B-other ALL.
Aims
The aim of this study was to screen UKALL14 patients with B-other ALL for ABL-class (ABL1, ABL2, PDGFRB, CSF1R), JAK-STAT (CRLF2, JAK2), ZNF384, and MEF2D rearrangements and determine their outcome to assess whether or not future such patients should be re-classified as HR.
Methods
Fixed cells left-over from diagnostic marrow samples were screened using commercially available and/or custom made dual colour break apart or dual fusion probes to detect gene rearrangements using standard methods. Endpoints were defined as per the trial protocol and analysed using standard methods. Patients were followed-up for a median of 2 years.
Results
Among 648 patients with B-cell precursor (BCP) ALL treated on UKALL14 (ISRCTN 66541317), 329 (51%) had HR genetics: BCR-ABL1 (n=196), KMT2A-AFF1 (n=47), low hypodiploidy (n=51) or a complex karyotype (n=35). The remaining 319 (49%) patients were classified as having SR genetics. Forty-seven patients had specific SR abnormalities: high hyperdiploidy (n=17), t(1;19) (n=15), other KMT2A (n=10), iAMP21 (n=4), ETV6-RUNX1 (n=1)]. The remaining patients were classified as B-other (n=190) or had failed cytogenetics (n=82). Patients with B-other ALL, complex or failed cytogenetics were screened and rearrangements were detected at the following frequencies: ABL1 (3/294, 1.0%), ABL2 (0/185), PDGFRB (3/187, 1.6%), CSF1R (0/187), CRLF2 (24/188, 13%), JAK2 (3/190, 1.6%), ZNF384 (11/145, 7.6%) and MEF2D (3/137, 2.2%). The following partners have been confirmed to date: EBF1-PDGFRB (n=1), IGH-CRLF2 (n=19), P2RY8-CRLF2 (n=5), BCR-JAK2 (n=1), PAX5-JAK2 (n=1) and TCF3-ZNF384 (n=6). Among 19 cases with a complex karyotype, none harboured an ABL-class or JAK-STAT abnormality but three had ZNF384 (n=2) or MEF2D (n=1) rearrangements.
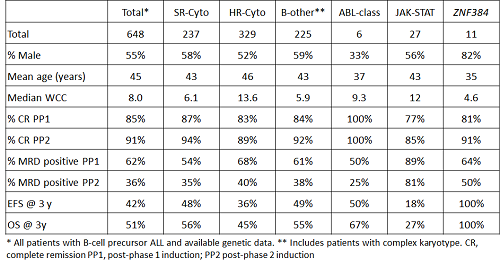
The table shows the demographic, clinical and outcome features of patients with ABL-class, JAK-STAT and ZNF384 abnormalities. Patients with ZNF384 or ABL-class abnormalities had a lower median age at diagnosis compared to other BCP-ALL cases (p=0.003 & p=0.06 respectively). Patients with JAK-STAT abnormalities had significantly inferior responses to initial treatment with higher rates of MRD positivity after phase 1 and phase 2 induction (p=0.015 & p<0.001, respectively). This poor response translated to a lower EFS and OS compared with other BCP-ALL patients (both p=0.01) due to higher rates of relapse/death and death: hazard ratio 1.74 (95% CI 1.10-2.73), p=0.017 and 1.81 (1.12-2.92), p=0.015, respectively. No patient with a ZNF384 abnormality suffered a relapse or death after a median follow-up time of 2 years (10 months to 4.9 years). EFS and OS rates for the 11 patients identified were significantly higher than other BCP-ALL patients (p=0.005 & p=0.014).
Conclusion
The frequency of ABL-class abnormalities was <1% in BCP-ALL. Approximately 5% of patients harboured a CRLF2 or JAK2 abnormality and collectively had a significantly inferior outcome. We suggest that patients with a JAK-STAT abnormality should be classified as high-risk. Finally, 11 patients with a ZNF384 fusion had 100% 3 years OS and EFS despite the lack of 100% MRD response.
Session topic: 1. Acute lymphoblastic leukemia – Biology & Translational Research
Keyword(s): Acute lymphoblastic leukemia, adult, Chromosomal abnormality, Prognostic factor
Abstract: S826
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 12:15 - 12:30
Location: Room A6
Background
Chromosomal abnormalities are established prognostic markers in adult ALL. In the UKALL14 clinical trial patients with BCR-ABL1, KMT2A-AFF1, low hypodiploidy or a complex karyotype were classified as having high risk (HR) genetics. All remaining patients, including those lacking an established chromosomal abnormality (B-other ALL) were classified as having standard risk (HR) genetics. Recent genomic studies have identified a plethora of new gene fusions within B-other ALL.
Aims
The aim of this study was to screen UKALL14 patients with B-other ALL for ABL-class (ABL1, ABL2, PDGFRB, CSF1R), JAK-STAT (CRLF2, JAK2), ZNF384, and MEF2D rearrangements and determine their outcome to assess whether or not future such patients should be re-classified as HR.
Methods
Fixed cells left-over from diagnostic marrow samples were screened using commercially available and/or custom made dual colour break apart or dual fusion probes to detect gene rearrangements using standard methods. Endpoints were defined as per the trial protocol and analysed using standard methods. Patients were followed-up for a median of 2 years.
Results
Among 648 patients with B-cell precursor (BCP) ALL treated on UKALL14 (ISRCTN 66541317), 329 (51%) had HR genetics: BCR-ABL1 (n=196), KMT2A-AFF1 (n=47), low hypodiploidy (n=51) or a complex karyotype (n=35). The remaining 319 (49%) patients were classified as having SR genetics. Forty-seven patients had specific SR abnormalities: high hyperdiploidy (n=17), t(1;19) (n=15), other KMT2A (n=10), iAMP21 (n=4), ETV6-RUNX1 (n=1)]. The remaining patients were classified as B-other (n=190) or had failed cytogenetics (n=82). Patients with B-other ALL, complex or failed cytogenetics were screened and rearrangements were detected at the following frequencies: ABL1 (3/294, 1.0%), ABL2 (0/185), PDGFRB (3/187, 1.6%), CSF1R (0/187), CRLF2 (24/188, 13%), JAK2 (3/190, 1.6%), ZNF384 (11/145, 7.6%) and MEF2D (3/137, 2.2%). The following partners have been confirmed to date: EBF1-PDGFRB (n=1), IGH-CRLF2 (n=19), P2RY8-CRLF2 (n=5), BCR-JAK2 (n=1), PAX5-JAK2 (n=1) and TCF3-ZNF384 (n=6). Among 19 cases with a complex karyotype, none harboured an ABL-class or JAK-STAT abnormality but three had ZNF384 (n=2) or MEF2D (n=1) rearrangements.
The table shows the demographic, clinical and outcome features of patients with ABL-class, JAK-STAT and ZNF384 abnormalities. Patients with ZNF384 or ABL-class abnormalities had a lower median age at diagnosis compared to other BCP-ALL cases (p=0.003 & p=0.06 respectively). Patients with JAK-STAT abnormalities had significantly inferior responses to initial treatment with higher rates of MRD positivity after phase 1 and phase 2 induction (p=0.015 & p<0.001, respectively). This poor response translated to a lower EFS and OS compared with other BCP-ALL patients (both p=0.01) due to higher rates of relapse/death and death: hazard ratio 1.74 (95% CI 1.10-2.73), p=0.017 and 1.81 (1.12-2.92), p=0.015, respectively. No patient with a ZNF384 abnormality suffered a relapse or death after a median follow-up time of 2 years (10 months to 4.9 years). EFS and OS rates for the 11 patients identified were significantly higher than other BCP-ALL patients (p=0.005 & p=0.014).

Conclusion
The frequency of ABL-class abnormalities was <1% in BCP-ALL. Approximately 5% of patients harboured a CRLF2 or JAK2 abnormality and collectively had a significantly inferior outcome. We suggest that patients with a JAK-STAT abnormality should be classified as high-risk. Finally, 11 patients with a ZNF384 fusion had 100% 3 years OS and EFS despite the lack of 100% MRD response.
Session topic: 1. Acute lymphoblastic leukemia – Biology & Translational Research
Keyword(s): Acute lymphoblastic leukemia, adult, Chromosomal abnormality, Prognostic factor


