
Contributions
Abstract: PB1805
Type: Publication Only
Background
Conventional cytogenetics is a common modality for tyrosine kinase inhibitor (TKI) response assessment in chronic myelogenous leukemia (CML) patients. There is no consensus regarding the use of conventional bone marrow (BM) cytogenetics or peripheral blood (PB) interphase fluorescence in situ hybridization (I-FISH) during follow-up. The routine dual colour FISH probes are less sensitive to reliably identify der(9) deletions during follow-up. BCR/ABL/ASS1 tri-colour dual fusion (TCDF) probe is highly sensitive and specific in identifying der(9) deletions.
Aims
Our aim was to identify the I-FISH fusion patterns of BCR/ABL/ASS1 TCDF probe and correlate the patterns with patient-specific molecular genetic parameters.
Methods
Results
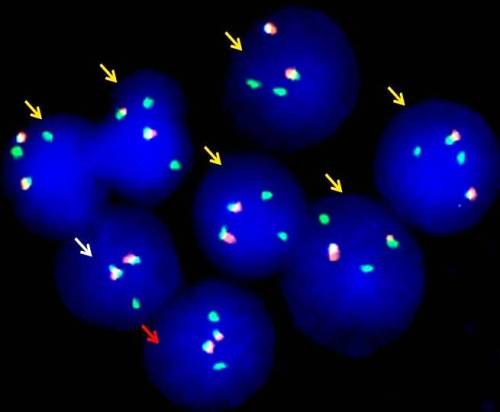
Conclusion
Our work would be an appropriate reference material for I-FISH signal interpretation using BCR/ABL/ASS1 TCDF probe. We have demonstrated a high frequency of der(9) deletions, clonal heterogeneity and absence of BCR-ABL1 amplification in an imatinib-resistant Indian CML cohort. For the first time, a significant association of der(9) deleted cell percentage with b2a2 transcript type and disease transformation status has been identified and the same has to be tested in a larger cohort.
Session topic: 7. Chronic myeloid leukemia - Biology
Keyword(s): Chronic myeloid leukemia, FISH
Abstract: PB1805
Type: Publication Only
Background
Conventional cytogenetics is a common modality for tyrosine kinase inhibitor (TKI) response assessment in chronic myelogenous leukemia (CML) patients. There is no consensus regarding the use of conventional bone marrow (BM) cytogenetics or peripheral blood (PB) interphase fluorescence in situ hybridization (I-FISH) during follow-up. The routine dual colour FISH probes are less sensitive to reliably identify der(9) deletions during follow-up. BCR/ABL/ASS1 tri-colour dual fusion (TCDF) probe is highly sensitive and specific in identifying der(9) deletions.
Aims
Our aim was to identify the I-FISH fusion patterns of BCR/ABL/ASS1 TCDF probe and correlate the patterns with patient-specific molecular genetic parameters.
Methods
Results

Conclusion
Our work would be an appropriate reference material for I-FISH signal interpretation using BCR/ABL/ASS1 TCDF probe. We have demonstrated a high frequency of der(9) deletions, clonal heterogeneity and absence of BCR-ABL1 amplification in an imatinib-resistant Indian CML cohort. For the first time, a significant association of der(9) deleted cell percentage with b2a2 transcript type and disease transformation status has been identified and the same has to be tested in a larger cohort.
Session topic: 7. Chronic myeloid leukemia - Biology
Keyword(s): Chronic myeloid leukemia, FISH


