
Contributions
Abstract: S491
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:00 - 16:15
Location: Room N103
Background
Clinical applications of next generation sequencing (NGS) in allogeneic hematopoietic stem cell transplantation are a topic of interest. Mutation dynamics post-HCT using longitudinal NGS have not been thoroughly examined. We hypothesized that serial sequencing of pre-HCT and post-HCT in AML patients could provide a much deeper and broader understanding of clonal origin/hierarchy of relapse after allogeneic HCT. The present study aimed to evaluate mutation dynamics in AML using serial samples from pre- and post-HCT with respect to transplant outcomes, particularly overall survival (OS) and relapse.
Aims
To track origins of post-HCT relapse in AML using serial sequencing
Methods
88 AML patients were enrolled and sequenced using an Illumina HiSeq 2000 sequencer (84 myeloid custom gene panel) on 419 bone marrow samples at diagnosis (n=88), pre-HCT (n=88), 21 days after HCT (n=88), and at relapse (n=20). Two patients relapsed by day 21. T-cell (n=80) and donor samples (n=57) were also sequenced. All computational and statistical analyses were performed using Python and R.
Results
The mean on-target coverage in 419 samples was 1773.7x. In total, we detected 217 mutations throughout the course of treatment in 79/88 patients (89.8%). NPM1 (26.1%), DNMT3A (26.1%), CEBPA (13.6%), IDH2 (13.6%), FLT3 (12.5%), and PTPN11 (11.4%) were commonly mutated at diagnosis. Unsurprisingly, most mutations appeared at initial diagnosis (200/217, 92.1%). Only 1, 2, and 14 mutations were acquired/selected at pre-HCT (0.5%), day 21 (0.9%), and relapse (6.5%), respectively. Most mutations were cleared at pre-HCT (mean mutation allele frequency (VAF) from 27.4% to 2.9%) and were further reduced after HCT (mean VAF from 2.9% to 0.7%) (Fig A).
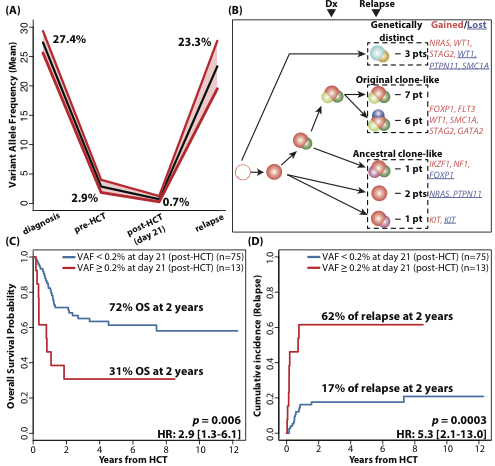
Conclusion
This study revealed origins of mutations detected at post-HCT relapse. It also revealed that the presence of mutations immediately after HCT, within 21 days, with low VAF (0.2%) can be used to predict relapse after HCT, illustrating the value of longitudinal NGS-based monitoring strategies for AML patients after allogeneic HCT.
Session topic: 22. Stem cell transplantation - Clinical
Keyword(s): AML, Relapsed acute myeloid leukemia, mutation analysis, Hematopoietic cell transplantation
Abstract: S491
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:00 - 16:15
Location: Room N103
Background
Clinical applications of next generation sequencing (NGS) in allogeneic hematopoietic stem cell transplantation are a topic of interest. Mutation dynamics post-HCT using longitudinal NGS have not been thoroughly examined. We hypothesized that serial sequencing of pre-HCT and post-HCT in AML patients could provide a much deeper and broader understanding of clonal origin/hierarchy of relapse after allogeneic HCT. The present study aimed to evaluate mutation dynamics in AML using serial samples from pre- and post-HCT with respect to transplant outcomes, particularly overall survival (OS) and relapse.
Aims
To track origins of post-HCT relapse in AML using serial sequencing
Methods
88 AML patients were enrolled and sequenced using an Illumina HiSeq 2000 sequencer (84 myeloid custom gene panel) on 419 bone marrow samples at diagnosis (n=88), pre-HCT (n=88), 21 days after HCT (n=88), and at relapse (n=20). Two patients relapsed by day 21. T-cell (n=80) and donor samples (n=57) were also sequenced. All computational and statistical analyses were performed using Python and R.
Results
The mean on-target coverage in 419 samples was 1773.7x. In total, we detected 217 mutations throughout the course of treatment in 79/88 patients (89.8%). NPM1 (26.1%), DNMT3A (26.1%), CEBPA (13.6%), IDH2 (13.6%), FLT3 (12.5%), and PTPN11 (11.4%) were commonly mutated at diagnosis. Unsurprisingly, most mutations appeared at initial diagnosis (200/217, 92.1%). Only 1, 2, and 14 mutations were acquired/selected at pre-HCT (0.5%), day 21 (0.9%), and relapse (6.5%), respectively. Most mutations were cleared at pre-HCT (mean mutation allele frequency (VAF) from 27.4% to 2.9%) and were further reduced after HCT (mean VAF from 2.9% to 0.7%) (Fig A).

Conclusion
This study revealed origins of mutations detected at post-HCT relapse. It also revealed that the presence of mutations immediately after HCT, within 21 days, with low VAF (0.2%) can be used to predict relapse after HCT, illustrating the value of longitudinal NGS-based monitoring strategies for AML patients after allogeneic HCT.
Session topic: 22. Stem cell transplantation - Clinical
Keyword(s): AML, Relapsed acute myeloid leukemia, mutation analysis, Hematopoietic cell transplantation


