
Contributions
Abstract: S486
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:00 - 16:15
Location: Room N105
Background
Cytopenia is a hallmark in myelodysplastic syndrome (MDS), however, many patients with persistent cytopenia do not fulfill the criteria for MDS. These patients are now classified as idiopathic cytopenia of undetermined significance (ICUS) or if a mutation is detected as clonal cytopenia of undetermined significance (CCUS). Little is known about these new entities in regards to survival and prognostication.
Aims
In this study we want to compare ICUS patients with MDS patients having low- or very low-risk disease according to the IPSS-R. We also wanted to investigate if sequencing of the cohort could bring additional information in regards to overall survival.
Methods
All patients underwent a bone marrow biopsy, cytogenetics and a broad range of blood tests. Furthermore, all ICUS patients underwent a blinded morphology review by two experienced pathologists; these review data will be ready for presentation at EHA. ICUS was defined as persistent cytopenia for more than six months, no chromosomal aberrations and common causes of cytopenia were ruled out. The patients were sequenced with a targeted sequencing panel, either using a customized Haloplex panel or a customized sequencing panel for the Ion Torrent platform. We analyzed 20 genes which are the most commonly mutated genes in MDS.
Results
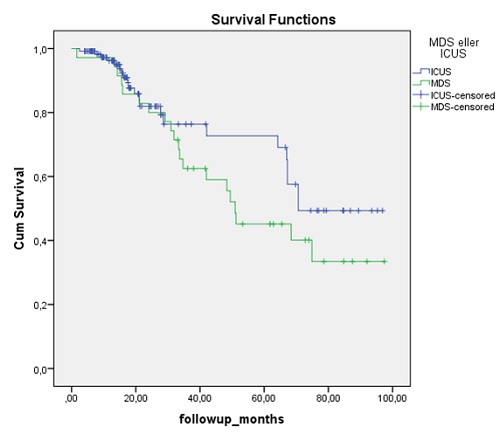
So far we included 157 patients, 122 were classified as ICUS and 35 as MDS and the median age is 65 and 68 years, respectively (p=0.27). We have sequenced 78% of the ICUS patients and 74% of the MDS patients. In total 53% and 73% of the ICUS and MDS patients had at least one mutation detected, respectively. If the patients carried a mutation, the median number of mutations was two in both the CCUS and the MDS group. The most commonly mutated genes were TET2, SRSF2, DNMT3A and ASXL1 in 38 patients (31%), n = 16 (13%), n = 10 (8%), n = 10 (8%), respectively. There were no significant differences in the distribution between the two groups. Mutations in NRAS, KRAS, TP53 were only identified in one patient each. The overall survival between the ICUS and the low-risk MDS patients did not differ (p=0.18) (figure 1). We also subdivided the ICUS patients into non-clonal ICUS and CCUS, but observed no difference between these two groups (p=0.355).
Conclusion
We here demonstrate that low-risk MDS and ICUS patients share similar survival patterns, however, larger studies with longer follow up are needed. Mutations are most commonly found in the epigenetic regulators in this cohort of ICUS and low-risk MDS, while mutations in classical tumor suppressors and oncogenes such as TP53 and NRAS are rare. Mutational screening seems promising in detecting patients at risk of progression, however, other biomarkers for prognostication are warranted.
Session topic: 10. Myelodysplastic syndromes - Clinical
Keyword(s): Myelodysplasia, Mutation, Clonality
Abstract: S486
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:00 - 16:15
Location: Room N105
Background
Cytopenia is a hallmark in myelodysplastic syndrome (MDS), however, many patients with persistent cytopenia do not fulfill the criteria for MDS. These patients are now classified as idiopathic cytopenia of undetermined significance (ICUS) or if a mutation is detected as clonal cytopenia of undetermined significance (CCUS). Little is known about these new entities in regards to survival and prognostication.
Aims
In this study we want to compare ICUS patients with MDS patients having low- or very low-risk disease according to the IPSS-R. We also wanted to investigate if sequencing of the cohort could bring additional information in regards to overall survival.
Methods
All patients underwent a bone marrow biopsy, cytogenetics and a broad range of blood tests. Furthermore, all ICUS patients underwent a blinded morphology review by two experienced pathologists; these review data will be ready for presentation at EHA. ICUS was defined as persistent cytopenia for more than six months, no chromosomal aberrations and common causes of cytopenia were ruled out. The patients were sequenced with a targeted sequencing panel, either using a customized Haloplex panel or a customized sequencing panel for the Ion Torrent platform. We analyzed 20 genes which are the most commonly mutated genes in MDS.
Results
So far we included 157 patients, 122 were classified as ICUS and 35 as MDS and the median age is 65 and 68 years, respectively (p=0.27). We have sequenced 78% of the ICUS patients and 74% of the MDS patients. In total 53% and 73% of the ICUS and MDS patients had at least one mutation detected, respectively. If the patients carried a mutation, the median number of mutations was two in both the CCUS and the MDS group. The most commonly mutated genes were TET2, SRSF2, DNMT3A and ASXL1 in 38 patients (31%), n = 16 (13%), n = 10 (8%), n = 10 (8%), respectively. There were no significant differences in the distribution between the two groups. Mutations in NRAS, KRAS, TP53 were only identified in one patient each. The overall survival between the ICUS and the low-risk MDS patients did not differ (p=0.18) (figure 1). We also subdivided the ICUS patients into non-clonal ICUS and CCUS, but observed no difference between these two groups (p=0.355).

Conclusion
We here demonstrate that low-risk MDS and ICUS patients share similar survival patterns, however, larger studies with longer follow up are needed. Mutations are most commonly found in the epigenetic regulators in this cohort of ICUS and low-risk MDS, while mutations in classical tumor suppressors and oncogenes such as TP53 and NRAS are rare. Mutational screening seems promising in detecting patients at risk of progression, however, other biomarkers for prognostication are warranted.
Session topic: 10. Myelodysplastic syndromes - Clinical
Keyword(s): Myelodysplasia, Mutation, Clonality


