SINGLE-CELL PROFILING OF HUMAN MEGAKARYOCYTE-ERYTHROID PROGENITORS IDENTIFIES DISTINCT MEGAKARYOCYTE AND ERYTHROID DIFFERENTIATION PATHWAYS
(Abstract release date: 05/19/16)
EHA Library. Psaila B. 06/10/16; 135175; S142

Dr. Beth Psaila
Contributions
Contributions
Abstract
Abstract: S142
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:45 - 12:00
Location: Room H5
Background
The conventional haematopoietic hierarchy proposes that megakaryocytes (MK) and erythroid cells (E) derive from a shared progenitor, the MEP. However, the MEP was defined using “bulk” assays and recent studies have suggested that MEP and other myeloid progenitor populations may be heterogeneous. Advances in single-cell techniques now provide the opportunity to dissect cellular heterogeneity within populations and to uncover rare cell types.
Aims
To integrate multiple different single-cell molecular and cell biology approaches to uncover heterogeneity in human MEP and to establish a new FACS strategy to prospectively purify and functionally validate the novel subpopulations.
Methods
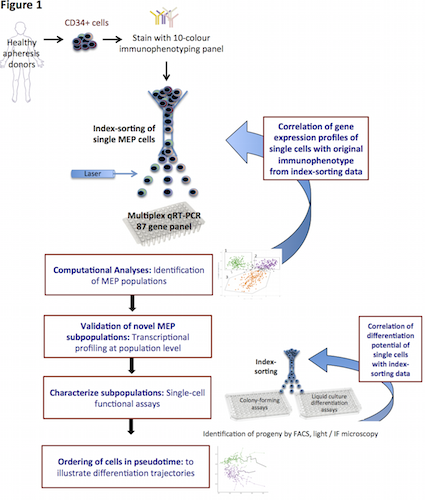
Surface immunophenotype (10 markers) and gene expression profiles (GEP) of individual human MEP were correlated using index-FACS and parallel, targeted multiplex PCR using a specifically designed panel of 87 genes. Differentiation potential was tested in novel single-cell differentiation assays (Figure).
Results
A total of 681 single Lin-CD34+CD38+CD123-CD45RA-MEPs were analysed by index sorting and GEP. Principal component analysis of GEP data identified three distinct MEP subpopulations. Correlation with cell surface immunophenotype revealed how these novel populations could be prospectively purified using the additional cell surface markers CD44, CD71 and CD41. MEP Population 1 (CD44hi71-41-; ~43.6% of MEP) showed increased expression of CD34, CSF3R and FLT3 and lower expression of GATA1 in single-cell and population-based analyses. Population 2 (CD44mod CD71+CD41-; 37.4%) showed increased E-associated gene expression (e.g. KLF1, TMOD1, TAL1, LEF1). Population 3 (CD44mod CD71+CD41+; 5.1%) showed distinct expression of MK-associated genes (e.g. FLI1, VWF, NH1B, MPL). Single-cell in vitro differential assays demonstrated that the transcriptional profiles corresponded to functional differences in lineage potential: Population 1 (designated Pre-MEPs) contained almost all of the residual myeloid potential within the MEP compartment and showed frequent MK-E bipotent cells. In contrast, populations 2 (E-MEP) and 3 (MK-MEP) showed a marked lineage bias toward E and MK lineage commitment respectively. Pseudo-temporal ordering of the cells along their differentiation trajectory (Monocle analysis) was also compatible with a more primitive phenotype for Pre-MEP, and suggested that CD42 expression correlated with loss of E-associated gene expression, a finding confirmed in functional assays with high MK but absence of E potential in CD71+CD41+CD42+ MEPs, while CD71+41+42- MK-MEP retained residual low level capacity to generate E progeny.
Conclusion
This report illustrates the power of combining three single-cell techniques to interrogate cellular heterogeneity within populations, an approach that is applicable to many other systems. We show that classically defined MEP in fact contain three distinct subpopulations: (1)“Pre-MEP”, enriched for E/MK progenitors but with residual myeloid differentiation capacity, (2)“E-MEP”, strongly biased towards E differentiation, and (3)“MK-MEP”, a rare population of bipotent cells that primarily generate MK progeny. Importantly, only a minority of classically defined MEP give rise to mixed E/MK colonies; the majority of the cells are transcriptionally-primed to generate cells of a single lineage. We believe that prospective identification of specific intermediate progenitor populations will allow for in-depth study of disorders of erythro-megakaryopoiesis, including myeloproliferative neoplasms and erythromegakaryocytic leukemias.
Session topic: Stem cells and the microenvironment
Keyword(s): Erythropoieisis, Hematopoiesis, Megakaryopoiesis, Thrombopoiesis
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:45 - 12:00
Location: Room H5
Background
The conventional haematopoietic hierarchy proposes that megakaryocytes (MK) and erythroid cells (E) derive from a shared progenitor, the MEP. However, the MEP was defined using “bulk” assays and recent studies have suggested that MEP and other myeloid progenitor populations may be heterogeneous. Advances in single-cell techniques now provide the opportunity to dissect cellular heterogeneity within populations and to uncover rare cell types.
Aims
To integrate multiple different single-cell molecular and cell biology approaches to uncover heterogeneity in human MEP and to establish a new FACS strategy to prospectively purify and functionally validate the novel subpopulations.
Methods
Surface immunophenotype (10 markers) and gene expression profiles (GEP) of individual human MEP were correlated using index-FACS and parallel, targeted multiplex PCR using a specifically designed panel of 87 genes. Differentiation potential was tested in novel single-cell differentiation assays (Figure).
Results
A total of 681 single Lin-CD34+CD38+CD123-CD45RA-MEPs were analysed by index sorting and GEP. Principal component analysis of GEP data identified three distinct MEP subpopulations. Correlation with cell surface immunophenotype revealed how these novel populations could be prospectively purified using the additional cell surface markers CD44, CD71 and CD41. MEP Population 1 (CD44hi71-41-; ~43.6% of MEP) showed increased expression of CD34, CSF3R and FLT3 and lower expression of GATA1 in single-cell and population-based analyses. Population 2 (CD44mod CD71+CD41-; 37.4%) showed increased E-associated gene expression (e.g. KLF1, TMOD1, TAL1, LEF1). Population 3 (CD44mod CD71+CD41+; 5.1%) showed distinct expression of MK-associated genes (e.g. FLI1, VWF, NH1B, MPL). Single-cell in vitro differential assays demonstrated that the transcriptional profiles corresponded to functional differences in lineage potential: Population 1 (designated Pre-MEPs) contained almost all of the residual myeloid potential within the MEP compartment and showed frequent MK-E bipotent cells. In contrast, populations 2 (E-MEP) and 3 (MK-MEP) showed a marked lineage bias toward E and MK lineage commitment respectively. Pseudo-temporal ordering of the cells along their differentiation trajectory (Monocle analysis) was also compatible with a more primitive phenotype for Pre-MEP, and suggested that CD42 expression correlated with loss of E-associated gene expression, a finding confirmed in functional assays with high MK but absence of E potential in CD71+CD41+CD42+ MEPs, while CD71+41+42- MK-MEP retained residual low level capacity to generate E progeny.
Conclusion
This report illustrates the power of combining three single-cell techniques to interrogate cellular heterogeneity within populations, an approach that is applicable to many other systems. We show that classically defined MEP in fact contain three distinct subpopulations: (1)“Pre-MEP”, enriched for E/MK progenitors but with residual myeloid differentiation capacity, (2)“E-MEP”, strongly biased towards E differentiation, and (3)“MK-MEP”, a rare population of bipotent cells that primarily generate MK progeny. Importantly, only a minority of classically defined MEP give rise to mixed E/MK colonies; the majority of the cells are transcriptionally-primed to generate cells of a single lineage. We believe that prospective identification of specific intermediate progenitor populations will allow for in-depth study of disorders of erythro-megakaryopoiesis, including myeloproliferative neoplasms and erythromegakaryocytic leukemias.

Session topic: Stem cells and the microenvironment
Keyword(s): Erythropoieisis, Hematopoiesis, Megakaryopoiesis, Thrombopoiesis
Abstract: S142
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:45 - 12:00
Location: Room H5
Background
The conventional haematopoietic hierarchy proposes that megakaryocytes (MK) and erythroid cells (E) derive from a shared progenitor, the MEP. However, the MEP was defined using “bulk” assays and recent studies have suggested that MEP and other myeloid progenitor populations may be heterogeneous. Advances in single-cell techniques now provide the opportunity to dissect cellular heterogeneity within populations and to uncover rare cell types.
Aims
To integrate multiple different single-cell molecular and cell biology approaches to uncover heterogeneity in human MEP and to establish a new FACS strategy to prospectively purify and functionally validate the novel subpopulations.
Methods
Surface immunophenotype (10 markers) and gene expression profiles (GEP) of individual human MEP were correlated using index-FACS and parallel, targeted multiplex PCR using a specifically designed panel of 87 genes. Differentiation potential was tested in novel single-cell differentiation assays (Figure).
Results
A total of 681 single Lin-CD34+CD38+CD123-CD45RA-MEPs were analysed by index sorting and GEP. Principal component analysis of GEP data identified three distinct MEP subpopulations. Correlation with cell surface immunophenotype revealed how these novel populations could be prospectively purified using the additional cell surface markers CD44, CD71 and CD41. MEP Population 1 (CD44hi71-41-; ~43.6% of MEP) showed increased expression of CD34, CSF3R and FLT3 and lower expression of GATA1 in single-cell and population-based analyses. Population 2 (CD44mod CD71+CD41-; 37.4%) showed increased E-associated gene expression (e.g. KLF1, TMOD1, TAL1, LEF1). Population 3 (CD44mod CD71+CD41+; 5.1%) showed distinct expression of MK-associated genes (e.g. FLI1, VWF, NH1B, MPL). Single-cell in vitro differential assays demonstrated that the transcriptional profiles corresponded to functional differences in lineage potential: Population 1 (designated Pre-MEPs) contained almost all of the residual myeloid potential within the MEP compartment and showed frequent MK-E bipotent cells. In contrast, populations 2 (E-MEP) and 3 (MK-MEP) showed a marked lineage bias toward E and MK lineage commitment respectively. Pseudo-temporal ordering of the cells along their differentiation trajectory (Monocle analysis) was also compatible with a more primitive phenotype for Pre-MEP, and suggested that CD42 expression correlated with loss of E-associated gene expression, a finding confirmed in functional assays with high MK but absence of E potential in CD71+CD41+CD42+ MEPs, while CD71+41+42- MK-MEP retained residual low level capacity to generate E progeny.
Conclusion
This report illustrates the power of combining three single-cell techniques to interrogate cellular heterogeneity within populations, an approach that is applicable to many other systems. We show that classically defined MEP in fact contain three distinct subpopulations: (1)“Pre-MEP”, enriched for E/MK progenitors but with residual myeloid differentiation capacity, (2)“E-MEP”, strongly biased towards E differentiation, and (3)“MK-MEP”, a rare population of bipotent cells that primarily generate MK progeny. Importantly, only a minority of classically defined MEP give rise to mixed E/MK colonies; the majority of the cells are transcriptionally-primed to generate cells of a single lineage. We believe that prospective identification of specific intermediate progenitor populations will allow for in-depth study of disorders of erythro-megakaryopoiesis, including myeloproliferative neoplasms and erythromegakaryocytic leukemias.

Session topic: Stem cells and the microenvironment
Keyword(s): Erythropoieisis, Hematopoiesis, Megakaryopoiesis, Thrombopoiesis
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:45 - 12:00
Location: Room H5
Background
The conventional haematopoietic hierarchy proposes that megakaryocytes (MK) and erythroid cells (E) derive from a shared progenitor, the MEP. However, the MEP was defined using “bulk” assays and recent studies have suggested that MEP and other myeloid progenitor populations may be heterogeneous. Advances in single-cell techniques now provide the opportunity to dissect cellular heterogeneity within populations and to uncover rare cell types.
Aims
To integrate multiple different single-cell molecular and cell biology approaches to uncover heterogeneity in human MEP and to establish a new FACS strategy to prospectively purify and functionally validate the novel subpopulations.
Methods
Surface immunophenotype (10 markers) and gene expression profiles (GEP) of individual human MEP were correlated using index-FACS and parallel, targeted multiplex PCR using a specifically designed panel of 87 genes. Differentiation potential was tested in novel single-cell differentiation assays (Figure).
Results
A total of 681 single Lin-CD34+CD38+CD123-CD45RA-MEPs were analysed by index sorting and GEP. Principal component analysis of GEP data identified three distinct MEP subpopulations. Correlation with cell surface immunophenotype revealed how these novel populations could be prospectively purified using the additional cell surface markers CD44, CD71 and CD41. MEP Population 1 (CD44hi71-41-; ~43.6% of MEP) showed increased expression of CD34, CSF3R and FLT3 and lower expression of GATA1 in single-cell and population-based analyses. Population 2 (CD44mod CD71+CD41-; 37.4%) showed increased E-associated gene expression (e.g. KLF1, TMOD1, TAL1, LEF1). Population 3 (CD44mod CD71+CD41+; 5.1%) showed distinct expression of MK-associated genes (e.g. FLI1, VWF, NH1B, MPL). Single-cell in vitro differential assays demonstrated that the transcriptional profiles corresponded to functional differences in lineage potential: Population 1 (designated Pre-MEPs) contained almost all of the residual myeloid potential within the MEP compartment and showed frequent MK-E bipotent cells. In contrast, populations 2 (E-MEP) and 3 (MK-MEP) showed a marked lineage bias toward E and MK lineage commitment respectively. Pseudo-temporal ordering of the cells along their differentiation trajectory (Monocle analysis) was also compatible with a more primitive phenotype for Pre-MEP, and suggested that CD42 expression correlated with loss of E-associated gene expression, a finding confirmed in functional assays with high MK but absence of E potential in CD71+CD41+CD42+ MEPs, while CD71+41+42- MK-MEP retained residual low level capacity to generate E progeny.
Conclusion
This report illustrates the power of combining three single-cell techniques to interrogate cellular heterogeneity within populations, an approach that is applicable to many other systems. We show that classically defined MEP in fact contain three distinct subpopulations: (1)“Pre-MEP”, enriched for E/MK progenitors but with residual myeloid differentiation capacity, (2)“E-MEP”, strongly biased towards E differentiation, and (3)“MK-MEP”, a rare population of bipotent cells that primarily generate MK progeny. Importantly, only a minority of classically defined MEP give rise to mixed E/MK colonies; the majority of the cells are transcriptionally-primed to generate cells of a single lineage. We believe that prospective identification of specific intermediate progenitor populations will allow for in-depth study of disorders of erythro-megakaryopoiesis, including myeloproliferative neoplasms and erythromegakaryocytic leukemias.

Session topic: Stem cells and the microenvironment
Keyword(s): Erythropoieisis, Hematopoiesis, Megakaryopoiesis, Thrombopoiesis
{{ help_message }}
{{filter}}


