FREQUENT RECURRING MUTATIONS DISRUPT THE ANTI-PROLIFERATIVE FUNCTION OF ZBTB7AIN ACUTE MYELOID LEUKEMIA WITH T(8;21) TRANSLOCATION
(Abstract release date: 05/19/16)
EHA Library. Hartmann L. 06/10/16; 135152; S119

Mrs. Luise Hartmann
Contributions
Contributions
Abstract
Abstract: S119
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:30 - 11:45
Location: Hall A3
Background
The t(8;21)(q22;q22) translocation results in the RUNX1/RUNX1T1 rearrangement and is one of the most frequent chromosomal aberrations in AML. However, in vivo models indicate the requirement of additional lesions, such as of KIT or FLT3 mutations, for leukemogenesis as the RUNX1/RUNX1T1 fusion gene alone is not sufficient to induce leukemia.
Aims
We set out to identify cooperating mutations in AML patients with t(8;21) translocation.
Methods
Exome and targeted sequencing, DNA pull-down, transcription assay, immunofluorescence, retroviral transduction, gene expression profiling.
Results
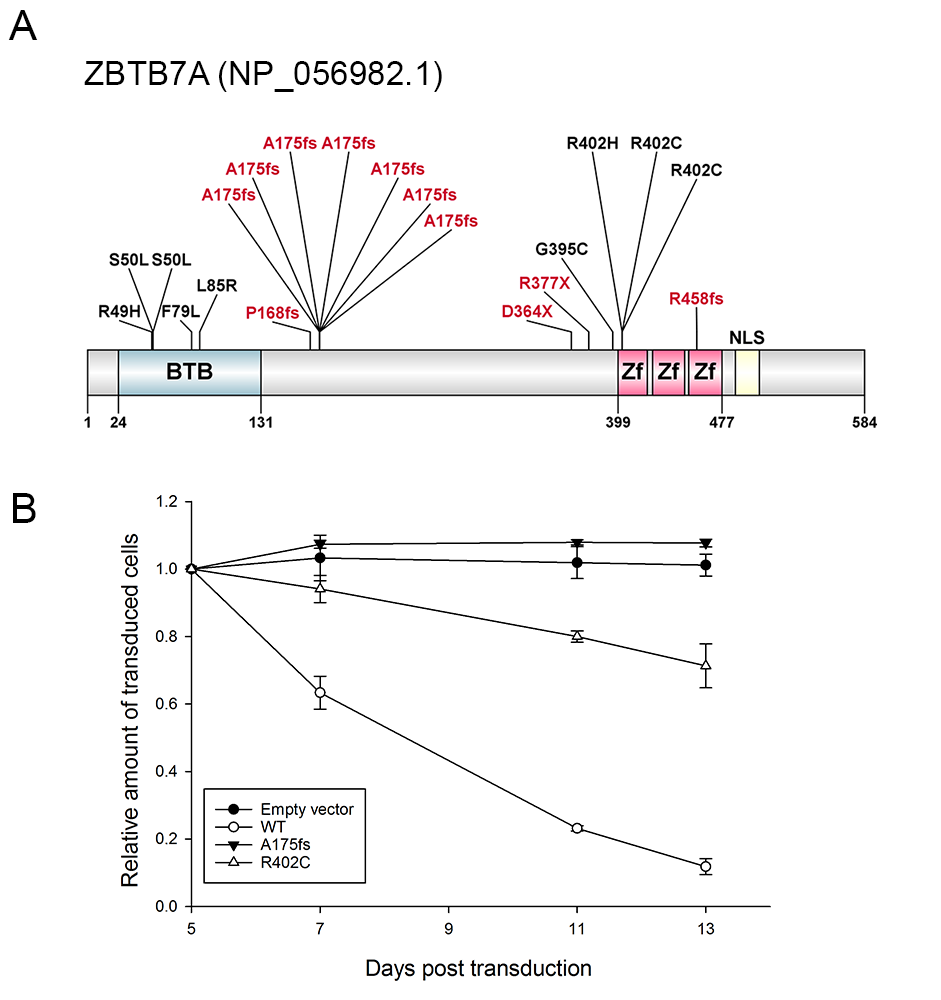
To identify additional cooperating mutations, we performed exome sequencing of matched diagnostic and remission samples from two AML patients with t(8;21) translocation. ZBTB7A was the only mutated gene identified in both patients. ZBTB7A is a member of the POZ/BTB and Krüppel (POK) transcription factor family. Previous studies suggested that ZBTB7A may act both as proto-oncogene and as tumor suppressor.Using targeted amplicon sequencing of ZBTB7A and 45 leukemia relevant genes, we screened 56 diagnostic AML t(8;21) samples. ZBTB7A mutations were identified in 13 of 56 patients (23%). Two recurring mutational hotspots (R402 and A175fs) in exon 2 were identified (Figure A). Variant allele frequency (VAF) ranged from 5.4-76.2% and 4 of 13 patients (31%) harbored two mutations of ZBTB7A. On the protein level, we confirmed the expression of a truncated ZBTB7A mutant (R377X) by Western blot for one patient with available material. In our cohort, ZBTB7A and ASXL2 mutations occurred at similar frequencies and 5 of 13 patients carried mutations in both genes, however, there was no significant association of mutated ZBTB7A and mutations in ASXL2 (Fisher’s exact test, p=0.12) or any other recurrently mutated gene.On a functional level, the ZBTB7A mutants R402H, R402C, A175fs or R377X failed to repress a luciferase reporter containing ZBTB7A-binding elements derived from the ARF-promoter. Structural modeling revealed that Arginine 402 binds into the major groove of the DNA double helix and likely contributes to the affinity or sequence specificity of the DNA interaction of the zinc finger domain of ZBTB7A. Through DNA pull-down assays we confirmed impaired DNA binding of A175fs and R402H. The truncating ZBTB7A mutants A175fs and R377X showed altered cytoplasmic protein localization. In the t(8;21) translocation positive AML cell line Kasumi-1 retroviral expression of wild-type ZBTB7A inhibited cell growth, whereas this anti-proliferative effect was not observed upon expression of the A175fs ZBTB7A mutant . The R402C mutant expressing Kasumi1-cells showed a trend towards reduced cell growth, suggesting residual activity (Figure B). Based on this observation, we expressed ZBTB7A wild-type or its mutants together with the RUNX1/RUNX1T1 fusion in lineage negative murine bone marrow cells and performed colony forming cell (CFC) assays. ZBTB7A expression led to a significant decrease in the number of colonies in primary CFC (87±12.6 versus 45±5.8, p<0.0001), while this effect was lost for both mutants A175fs and R402C. These findings support an oncogenic synergism between RUNX1/RUNX1T1 and ZBTB7A mutations.We correlated ZBTB7A expression with clinical outcome in a large cohort of AML patients (GSE37642). Remarkably, in over 200 cytogenetically normal AML patients treated on clinical trial (NCT00266136), high expression of ZBTB7A was associated with a favorable outcome (p=0.0004), suggesting a relevance in AML beyond the t(8;21) subgroup.
Conclusion
In summary, we have identified ZBTB7A as one of the most frequently mutated genes in t(8;21) positive AML. Considering that ZBTB7A mutations result in loss of function, we suggest that ZBTB7A acts as a tumor suppressor in t(8;21) positive AML.
Session topic: AML Biology - Novel mechanisms of leukemogenesis
Keyword(s): Acute myeloid leukemia, Mutation analysis
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:30 - 11:45
Location: Hall A3
Background
The t(8;21)(q22;q22) translocation results in the RUNX1/RUNX1T1 rearrangement and is one of the most frequent chromosomal aberrations in AML. However, in vivo models indicate the requirement of additional lesions, such as of KIT or FLT3 mutations, for leukemogenesis as the RUNX1/RUNX1T1 fusion gene alone is not sufficient to induce leukemia.
Aims
We set out to identify cooperating mutations in AML patients with t(8;21) translocation.
Methods
Exome and targeted sequencing, DNA pull-down, transcription assay, immunofluorescence, retroviral transduction, gene expression profiling.
Results
To identify additional cooperating mutations, we performed exome sequencing of matched diagnostic and remission samples from two AML patients with t(8;21) translocation. ZBTB7A was the only mutated gene identified in both patients. ZBTB7A is a member of the POZ/BTB and Krüppel (POK) transcription factor family. Previous studies suggested that ZBTB7A may act both as proto-oncogene and as tumor suppressor.Using targeted amplicon sequencing of ZBTB7A and 45 leukemia relevant genes, we screened 56 diagnostic AML t(8;21) samples. ZBTB7A mutations were identified in 13 of 56 patients (23%). Two recurring mutational hotspots (R402 and A175fs) in exon 2 were identified (Figure A). Variant allele frequency (VAF) ranged from 5.4-76.2% and 4 of 13 patients (31%) harbored two mutations of ZBTB7A. On the protein level, we confirmed the expression of a truncated ZBTB7A mutant (R377X) by Western blot for one patient with available material. In our cohort, ZBTB7A and ASXL2 mutations occurred at similar frequencies and 5 of 13 patients carried mutations in both genes, however, there was no significant association of mutated ZBTB7A and mutations in ASXL2 (Fisher’s exact test, p=0.12) or any other recurrently mutated gene.On a functional level, the ZBTB7A mutants R402H, R402C, A175fs or R377X failed to repress a luciferase reporter containing ZBTB7A-binding elements derived from the ARF-promoter. Structural modeling revealed that Arginine 402 binds into the major groove of the DNA double helix and likely contributes to the affinity or sequence specificity of the DNA interaction of the zinc finger domain of ZBTB7A. Through DNA pull-down assays we confirmed impaired DNA binding of A175fs and R402H. The truncating ZBTB7A mutants A175fs and R377X showed altered cytoplasmic protein localization. In the t(8;21) translocation positive AML cell line Kasumi-1 retroviral expression of wild-type ZBTB7A inhibited cell growth, whereas this anti-proliferative effect was not observed upon expression of the A175fs ZBTB7A mutant . The R402C mutant expressing Kasumi1-cells showed a trend towards reduced cell growth, suggesting residual activity (Figure B). Based on this observation, we expressed ZBTB7A wild-type or its mutants together with the RUNX1/RUNX1T1 fusion in lineage negative murine bone marrow cells and performed colony forming cell (CFC) assays. ZBTB7A expression led to a significant decrease in the number of colonies in primary CFC (87±12.6 versus 45±5.8, p<0.0001), while this effect was lost for both mutants A175fs and R402C. These findings support an oncogenic synergism between RUNX1/RUNX1T1 and ZBTB7A mutations.We correlated ZBTB7A expression with clinical outcome in a large cohort of AML patients (GSE37642). Remarkably, in over 200 cytogenetically normal AML patients treated on clinical trial (NCT00266136), high expression of ZBTB7A was associated with a favorable outcome (p=0.0004), suggesting a relevance in AML beyond the t(8;21) subgroup.
Conclusion
In summary, we have identified ZBTB7A as one of the most frequently mutated genes in t(8;21) positive AML. Considering that ZBTB7A mutations result in loss of function, we suggest that ZBTB7A acts as a tumor suppressor in t(8;21) positive AML.

Session topic: AML Biology - Novel mechanisms of leukemogenesis
Keyword(s): Acute myeloid leukemia, Mutation analysis
Abstract: S119
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:30 - 11:45
Location: Hall A3
Background
The t(8;21)(q22;q22) translocation results in the RUNX1/RUNX1T1 rearrangement and is one of the most frequent chromosomal aberrations in AML. However, in vivo models indicate the requirement of additional lesions, such as of KIT or FLT3 mutations, for leukemogenesis as the RUNX1/RUNX1T1 fusion gene alone is not sufficient to induce leukemia.
Aims
We set out to identify cooperating mutations in AML patients with t(8;21) translocation.
Methods
Exome and targeted sequencing, DNA pull-down, transcription assay, immunofluorescence, retroviral transduction, gene expression profiling.
Results
To identify additional cooperating mutations, we performed exome sequencing of matched diagnostic and remission samples from two AML patients with t(8;21) translocation. ZBTB7A was the only mutated gene identified in both patients. ZBTB7A is a member of the POZ/BTB and Krüppel (POK) transcription factor family. Previous studies suggested that ZBTB7A may act both as proto-oncogene and as tumor suppressor.Using targeted amplicon sequencing of ZBTB7A and 45 leukemia relevant genes, we screened 56 diagnostic AML t(8;21) samples. ZBTB7A mutations were identified in 13 of 56 patients (23%). Two recurring mutational hotspots (R402 and A175fs) in exon 2 were identified (Figure A). Variant allele frequency (VAF) ranged from 5.4-76.2% and 4 of 13 patients (31%) harbored two mutations of ZBTB7A. On the protein level, we confirmed the expression of a truncated ZBTB7A mutant (R377X) by Western blot for one patient with available material. In our cohort, ZBTB7A and ASXL2 mutations occurred at similar frequencies and 5 of 13 patients carried mutations in both genes, however, there was no significant association of mutated ZBTB7A and mutations in ASXL2 (Fisher’s exact test, p=0.12) or any other recurrently mutated gene.On a functional level, the ZBTB7A mutants R402H, R402C, A175fs or R377X failed to repress a luciferase reporter containing ZBTB7A-binding elements derived from the ARF-promoter. Structural modeling revealed that Arginine 402 binds into the major groove of the DNA double helix and likely contributes to the affinity or sequence specificity of the DNA interaction of the zinc finger domain of ZBTB7A. Through DNA pull-down assays we confirmed impaired DNA binding of A175fs and R402H. The truncating ZBTB7A mutants A175fs and R377X showed altered cytoplasmic protein localization. In the t(8;21) translocation positive AML cell line Kasumi-1 retroviral expression of wild-type ZBTB7A inhibited cell growth, whereas this anti-proliferative effect was not observed upon expression of the A175fs ZBTB7A mutant . The R402C mutant expressing Kasumi1-cells showed a trend towards reduced cell growth, suggesting residual activity (Figure B). Based on this observation, we expressed ZBTB7A wild-type or its mutants together with the RUNX1/RUNX1T1 fusion in lineage negative murine bone marrow cells and performed colony forming cell (CFC) assays. ZBTB7A expression led to a significant decrease in the number of colonies in primary CFC (87±12.6 versus 45±5.8, p<0.0001), while this effect was lost for both mutants A175fs and R402C. These findings support an oncogenic synergism between RUNX1/RUNX1T1 and ZBTB7A mutations.We correlated ZBTB7A expression with clinical outcome in a large cohort of AML patients (GSE37642). Remarkably, in over 200 cytogenetically normal AML patients treated on clinical trial (NCT00266136), high expression of ZBTB7A was associated with a favorable outcome (p=0.0004), suggesting a relevance in AML beyond the t(8;21) subgroup.
Conclusion
In summary, we have identified ZBTB7A as one of the most frequently mutated genes in t(8;21) positive AML. Considering that ZBTB7A mutations result in loss of function, we suggest that ZBTB7A acts as a tumor suppressor in t(8;21) positive AML.

Session topic: AML Biology - Novel mechanisms of leukemogenesis
Keyword(s): Acute myeloid leukemia, Mutation analysis
Type: Oral Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 11:30 - 11:45
Location: Hall A3
Background
The t(8;21)(q22;q22) translocation results in the RUNX1/RUNX1T1 rearrangement and is one of the most frequent chromosomal aberrations in AML. However, in vivo models indicate the requirement of additional lesions, such as of KIT or FLT3 mutations, for leukemogenesis as the RUNX1/RUNX1T1 fusion gene alone is not sufficient to induce leukemia.
Aims
We set out to identify cooperating mutations in AML patients with t(8;21) translocation.
Methods
Exome and targeted sequencing, DNA pull-down, transcription assay, immunofluorescence, retroviral transduction, gene expression profiling.
Results
To identify additional cooperating mutations, we performed exome sequencing of matched diagnostic and remission samples from two AML patients with t(8;21) translocation. ZBTB7A was the only mutated gene identified in both patients. ZBTB7A is a member of the POZ/BTB and Krüppel (POK) transcription factor family. Previous studies suggested that ZBTB7A may act both as proto-oncogene and as tumor suppressor.Using targeted amplicon sequencing of ZBTB7A and 45 leukemia relevant genes, we screened 56 diagnostic AML t(8;21) samples. ZBTB7A mutations were identified in 13 of 56 patients (23%). Two recurring mutational hotspots (R402 and A175fs) in exon 2 were identified (Figure A). Variant allele frequency (VAF) ranged from 5.4-76.2% and 4 of 13 patients (31%) harbored two mutations of ZBTB7A. On the protein level, we confirmed the expression of a truncated ZBTB7A mutant (R377X) by Western blot for one patient with available material. In our cohort, ZBTB7A and ASXL2 mutations occurred at similar frequencies and 5 of 13 patients carried mutations in both genes, however, there was no significant association of mutated ZBTB7A and mutations in ASXL2 (Fisher’s exact test, p=0.12) or any other recurrently mutated gene.On a functional level, the ZBTB7A mutants R402H, R402C, A175fs or R377X failed to repress a luciferase reporter containing ZBTB7A-binding elements derived from the ARF-promoter. Structural modeling revealed that Arginine 402 binds into the major groove of the DNA double helix and likely contributes to the affinity or sequence specificity of the DNA interaction of the zinc finger domain of ZBTB7A. Through DNA pull-down assays we confirmed impaired DNA binding of A175fs and R402H. The truncating ZBTB7A mutants A175fs and R377X showed altered cytoplasmic protein localization. In the t(8;21) translocation positive AML cell line Kasumi-1 retroviral expression of wild-type ZBTB7A inhibited cell growth, whereas this anti-proliferative effect was not observed upon expression of the A175fs ZBTB7A mutant . The R402C mutant expressing Kasumi1-cells showed a trend towards reduced cell growth, suggesting residual activity (Figure B). Based on this observation, we expressed ZBTB7A wild-type or its mutants together with the RUNX1/RUNX1T1 fusion in lineage negative murine bone marrow cells and performed colony forming cell (CFC) assays. ZBTB7A expression led to a significant decrease in the number of colonies in primary CFC (87±12.6 versus 45±5.8, p<0.0001), while this effect was lost for both mutants A175fs and R402C. These findings support an oncogenic synergism between RUNX1/RUNX1T1 and ZBTB7A mutations.We correlated ZBTB7A expression with clinical outcome in a large cohort of AML patients (GSE37642). Remarkably, in over 200 cytogenetically normal AML patients treated on clinical trial (NCT00266136), high expression of ZBTB7A was associated with a favorable outcome (p=0.0004), suggesting a relevance in AML beyond the t(8;21) subgroup.
Conclusion
In summary, we have identified ZBTB7A as one of the most frequently mutated genes in t(8;21) positive AML. Considering that ZBTB7A mutations result in loss of function, we suggest that ZBTB7A acts as a tumor suppressor in t(8;21) positive AML.

Session topic: AML Biology - Novel mechanisms of leukemogenesis
Keyword(s): Acute myeloid leukemia, Mutation analysis
{{ help_message }}
{{filter}}


