SERUM PROTEOMIC PROFILES OF HODGKIN?S LYMPHOMA PATIENTS SHOW DIFFERENTIAL EXPRESSION ACCORDING TO EPSTEIN-BARR VIRUS STATUS
(Abstract release date: 05/19/16)
EHA Library. Musacchio J. 06/09/16; 134946; PB2046

Dr. Juliane Musacchio
Contributions
Contributions
Abstract
Abstract: PB2046
Type: Publication Only
Background
Despite the advances in understanding of Hodgkin’s lymphoma’s molecular pathogenesis, its association with EBV remains unclear. In this study, we assessed serum protein profiles from EBV associated and non-associated Hodgkin’s lymphoma patients and identified three differentially expressed proteins that are candidate biomarkers.
Aims
The aim of the present study was to identify and analyze differentially expressed proteins in pooled sera from patients with HL according to EBV status.
Methods
Blood samples were obtained from 16 patients with recently diagnosed HL before treatment. Blood samples from 10 healthy volunteers were also included. The inclusion criteria used for EBV non-associated HL were the negativity of EBV in buffy coat, plasma, and lymph node. Conversely, patients with EBV associated HL should have EBV detected in buffy coat or plasma, and lymph node. MALDI-MS was performed using a 4700 Proteomics Analyzer® (Applied Biosystems).
Results
The patients’ median age at diagnosis was 26 years (range 15-56) and 38% were men. Our analysis revealed about 265 spots with a mean of 86 (35%) identified spots on each gel. Five spots were indicated as having different normalized volumes when compared to each other (p<0.05) (Figure1). Two up-regulated proteins were detected in patients with EBV non-associated HL, and identified as inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) and haptoglobin (HP). Only one protein, fibrinogen beta chain, was down-regulated in the serum from EBV-associated HL patients.
Conclusion
We identified three serum proteins that may serve as potential biomarkers in HL, according to EBV status.The biological function of ITIH4 is still unknown. A possible role is the modulation of cell migration and proliferation during the development of the acute-phase response. These findings may suggest that EBV presence could inhibit ITIH4 expression. Serum HP levels in the lymphoma patients have also been described to be significantly higher than in the control group and have been correlated to response to treatment. These results suggest HP as a new tumor marker for lymphoma. Here HP is not increased in patients with advanced stage disease, but in absence of systemic EBV infection, which is in line with prior observations: a recent study described some phenotypes with a higher HP concentration (HP 1-1 and HP 2-1) are less prone to positive EBV serology. In spite of FGB has been described as a serum acute phase reactant protein, their levels were decreased in patients diagnosed with EBV associated HL. The cytokines IL-4, IL-10 and IL-13 have a protective effect against vascular injury leading to atherosclerosis, dose dependently down regulate the biosynthesis of fibrinogen, and some of these cytokines are known involved in the HL pathophysiology. In conclusion, the present study represents an initial and necessary step needed to identify proteins that are differentially expressed between patients with EBV associated and non-associated HL. We demonstrated that protein profiling of serum significantly, accurately and reproducibility distinguished EBV associated and non-associated HL patients. This is the first report combining analysis of serum proteomics of HL patients, according to EBV status, and the pooled sera approach as an appropriate initial approach. Following the identification of spots, future studies with more samples are still required to validate our results. Besides such validation could be performed in large scale by other techniques (e.g., ELISA-based methods).
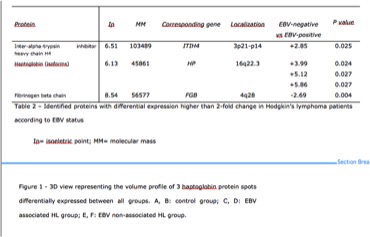
Session topic: E-poster
Keyword(s): EBV, Proteomics
Type: Publication Only
Background
Despite the advances in understanding of Hodgkin’s lymphoma’s molecular pathogenesis, its association with EBV remains unclear. In this study, we assessed serum protein profiles from EBV associated and non-associated Hodgkin’s lymphoma patients and identified three differentially expressed proteins that are candidate biomarkers.
Aims
The aim of the present study was to identify and analyze differentially expressed proteins in pooled sera from patients with HL according to EBV status.
Methods
Blood samples were obtained from 16 patients with recently diagnosed HL before treatment. Blood samples from 10 healthy volunteers were also included. The inclusion criteria used for EBV non-associated HL were the negativity of EBV in buffy coat, plasma, and lymph node. Conversely, patients with EBV associated HL should have EBV detected in buffy coat or plasma, and lymph node. MALDI-MS was performed using a 4700 Proteomics Analyzer® (Applied Biosystems).
Results
The patients’ median age at diagnosis was 26 years (range 15-56) and 38% were men. Our analysis revealed about 265 spots with a mean of 86 (35%) identified spots on each gel. Five spots were indicated as having different normalized volumes when compared to each other (p<0.05) (Figure1). Two up-regulated proteins were detected in patients with EBV non-associated HL, and identified as inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) and haptoglobin (HP). Only one protein, fibrinogen beta chain, was down-regulated in the serum from EBV-associated HL patients.
Conclusion
We identified three serum proteins that may serve as potential biomarkers in HL, according to EBV status.The biological function of ITIH4 is still unknown. A possible role is the modulation of cell migration and proliferation during the development of the acute-phase response. These findings may suggest that EBV presence could inhibit ITIH4 expression. Serum HP levels in the lymphoma patients have also been described to be significantly higher than in the control group and have been correlated to response to treatment. These results suggest HP as a new tumor marker for lymphoma. Here HP is not increased in patients with advanced stage disease, but in absence of systemic EBV infection, which is in line with prior observations: a recent study described some phenotypes with a higher HP concentration (HP 1-1 and HP 2-1) are less prone to positive EBV serology. In spite of FGB has been described as a serum acute phase reactant protein, their levels were decreased in patients diagnosed with EBV associated HL. The cytokines IL-4, IL-10 and IL-13 have a protective effect against vascular injury leading to atherosclerosis, dose dependently down regulate the biosynthesis of fibrinogen, and some of these cytokines are known involved in the HL pathophysiology. In conclusion, the present study represents an initial and necessary step needed to identify proteins that are differentially expressed between patients with EBV associated and non-associated HL. We demonstrated that protein profiling of serum significantly, accurately and reproducibility distinguished EBV associated and non-associated HL patients. This is the first report combining analysis of serum proteomics of HL patients, according to EBV status, and the pooled sera approach as an appropriate initial approach. Following the identification of spots, future studies with more samples are still required to validate our results. Besides such validation could be performed in large scale by other techniques (e.g., ELISA-based methods).

Session topic: E-poster
Keyword(s): EBV, Proteomics
Abstract: PB2046
Type: Publication Only
Background
Despite the advances in understanding of Hodgkin’s lymphoma’s molecular pathogenesis, its association with EBV remains unclear. In this study, we assessed serum protein profiles from EBV associated and non-associated Hodgkin’s lymphoma patients and identified three differentially expressed proteins that are candidate biomarkers.
Aims
The aim of the present study was to identify and analyze differentially expressed proteins in pooled sera from patients with HL according to EBV status.
Methods
Blood samples were obtained from 16 patients with recently diagnosed HL before treatment. Blood samples from 10 healthy volunteers were also included. The inclusion criteria used for EBV non-associated HL were the negativity of EBV in buffy coat, plasma, and lymph node. Conversely, patients with EBV associated HL should have EBV detected in buffy coat or plasma, and lymph node. MALDI-MS was performed using a 4700 Proteomics Analyzer® (Applied Biosystems).
Results
The patients’ median age at diagnosis was 26 years (range 15-56) and 38% were men. Our analysis revealed about 265 spots with a mean of 86 (35%) identified spots on each gel. Five spots were indicated as having different normalized volumes when compared to each other (p<0.05) (Figure1). Two up-regulated proteins were detected in patients with EBV non-associated HL, and identified as inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) and haptoglobin (HP). Only one protein, fibrinogen beta chain, was down-regulated in the serum from EBV-associated HL patients.
Conclusion
We identified three serum proteins that may serve as potential biomarkers in HL, according to EBV status.The biological function of ITIH4 is still unknown. A possible role is the modulation of cell migration and proliferation during the development of the acute-phase response. These findings may suggest that EBV presence could inhibit ITIH4 expression. Serum HP levels in the lymphoma patients have also been described to be significantly higher than in the control group and have been correlated to response to treatment. These results suggest HP as a new tumor marker for lymphoma. Here HP is not increased in patients with advanced stage disease, but in absence of systemic EBV infection, which is in line with prior observations: a recent study described some phenotypes with a higher HP concentration (HP 1-1 and HP 2-1) are less prone to positive EBV serology. In spite of FGB has been described as a serum acute phase reactant protein, their levels were decreased in patients diagnosed with EBV associated HL. The cytokines IL-4, IL-10 and IL-13 have a protective effect against vascular injury leading to atherosclerosis, dose dependently down regulate the biosynthesis of fibrinogen, and some of these cytokines are known involved in the HL pathophysiology. In conclusion, the present study represents an initial and necessary step needed to identify proteins that are differentially expressed between patients with EBV associated and non-associated HL. We demonstrated that protein profiling of serum significantly, accurately and reproducibility distinguished EBV associated and non-associated HL patients. This is the first report combining analysis of serum proteomics of HL patients, according to EBV status, and the pooled sera approach as an appropriate initial approach. Following the identification of spots, future studies with more samples are still required to validate our results. Besides such validation could be performed in large scale by other techniques (e.g., ELISA-based methods).

Session topic: E-poster
Keyword(s): EBV, Proteomics
Type: Publication Only
Background
Despite the advances in understanding of Hodgkin’s lymphoma’s molecular pathogenesis, its association with EBV remains unclear. In this study, we assessed serum protein profiles from EBV associated and non-associated Hodgkin’s lymphoma patients and identified three differentially expressed proteins that are candidate biomarkers.
Aims
The aim of the present study was to identify and analyze differentially expressed proteins in pooled sera from patients with HL according to EBV status.
Methods
Blood samples were obtained from 16 patients with recently diagnosed HL before treatment. Blood samples from 10 healthy volunteers were also included. The inclusion criteria used for EBV non-associated HL were the negativity of EBV in buffy coat, plasma, and lymph node. Conversely, patients with EBV associated HL should have EBV detected in buffy coat or plasma, and lymph node. MALDI-MS was performed using a 4700 Proteomics Analyzer® (Applied Biosystems).
Results
The patients’ median age at diagnosis was 26 years (range 15-56) and 38% were men. Our analysis revealed about 265 spots with a mean of 86 (35%) identified spots on each gel. Five spots were indicated as having different normalized volumes when compared to each other (p<0.05) (Figure1). Two up-regulated proteins were detected in patients with EBV non-associated HL, and identified as inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) and haptoglobin (HP). Only one protein, fibrinogen beta chain, was down-regulated in the serum from EBV-associated HL patients.
Conclusion
We identified three serum proteins that may serve as potential biomarkers in HL, according to EBV status.The biological function of ITIH4 is still unknown. A possible role is the modulation of cell migration and proliferation during the development of the acute-phase response. These findings may suggest that EBV presence could inhibit ITIH4 expression. Serum HP levels in the lymphoma patients have also been described to be significantly higher than in the control group and have been correlated to response to treatment. These results suggest HP as a new tumor marker for lymphoma. Here HP is not increased in patients with advanced stage disease, but in absence of systemic EBV infection, which is in line with prior observations: a recent study described some phenotypes with a higher HP concentration (HP 1-1 and HP 2-1) are less prone to positive EBV serology. In spite of FGB has been described as a serum acute phase reactant protein, their levels were decreased in patients diagnosed with EBV associated HL. The cytokines IL-4, IL-10 and IL-13 have a protective effect against vascular injury leading to atherosclerosis, dose dependently down regulate the biosynthesis of fibrinogen, and some of these cytokines are known involved in the HL pathophysiology. In conclusion, the present study represents an initial and necessary step needed to identify proteins that are differentially expressed between patients with EBV associated and non-associated HL. We demonstrated that protein profiling of serum significantly, accurately and reproducibility distinguished EBV associated and non-associated HL patients. This is the first report combining analysis of serum proteomics of HL patients, according to EBV status, and the pooled sera approach as an appropriate initial approach. Following the identification of spots, future studies with more samples are still required to validate our results. Besides such validation could be performed in large scale by other techniques (e.g., ELISA-based methods).

Session topic: E-poster
Keyword(s): EBV, Proteomics
{{ help_message }}
{{filter}}


