ASSESSMENT OF THE INTERLABORATORY VARIABILITY AND ROBUSTNESS OF JAK2V617F MUTATION ASSAYS: A STANDARDIZATION STUDY INVOLVING A CONSORTIUM OF 19 ITALIAN LABORATORIES
(Abstract release date: 05/19/16)
EHA Library. Perricone M. 06/09/16; 134909; PB2009

Mrs. Margherita Perricone
Contributions
Contributions
Abstract
Abstract: PB2009
Type: Publication Only
Background
In chronic myeloproliferative neoplasms (MPNs), the quantification of the JAK2V617F allele burden (AB) is crucial for diagnosis and prognosis assessment, and also for disease monitoring after allogeneic stem-cell transplantation. To-date, a plethora of techniques for JAK2V617F determination is used over different molecular laboratories, with substantial differences in specificity and sensitivity. Given the need to provide reliable and comparable molecular results, the standardization of molecular techniques is of utmost importance.
Aims
The aims of this multicenter study were: 1) to evaluate the inter- and intra-laboratory variability in JAK2V617F quantification in 19 Italian molecular laboratories; 2) to identify the most robust assay for the standardization of the molecular test; 3) to allow consistent interpretation of individual patient analysis results.
Methods
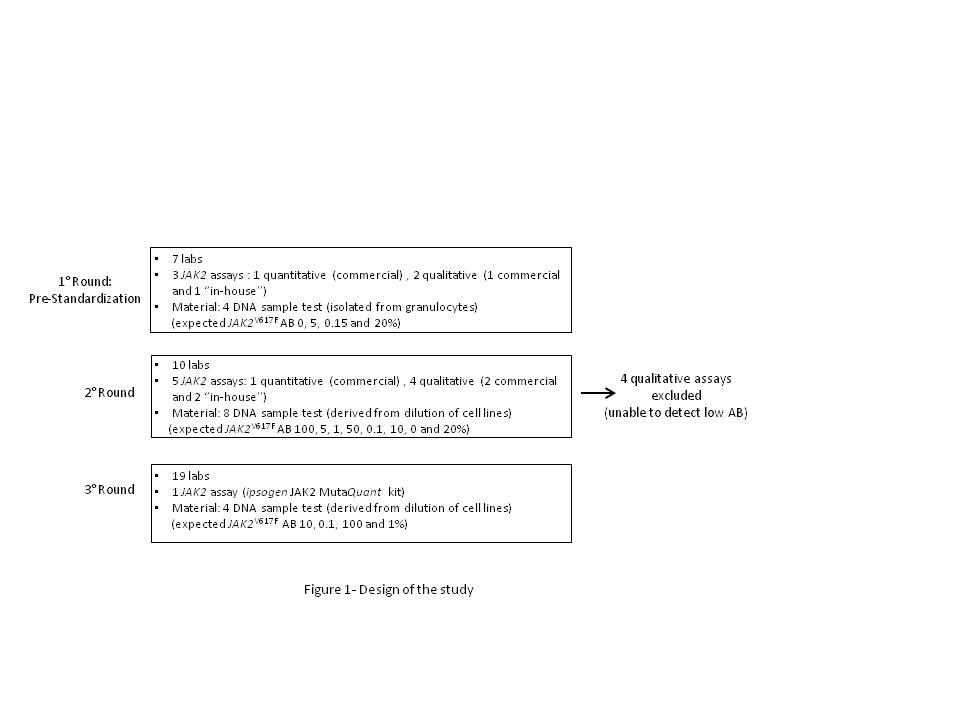
A network of 19 Italian molecular laboratories was established. The study was coordinated by the Institute of Hematology “L. and A. Seràgnoli”, Bologna, and was developed in 3 different rounds (Fig. 1). In routine practice, 2 laboratories did not assess JAK2V617F mutation whereas 7 used a qualitative approach and 10 performed a quantitative evaluation. Both reagents and DNA samples were provided by Werfen-IL SpA and QIAGEN. Raw data and runs validity were checked according to handbook recommendations. Statistical analysis was carried out by QIAGEN/Bologna University.
Results
In the 1st round, we aimed to investigate the inter-laboratory variability on different mutation loads. All laboratories using a quantitative approach were able to determine the expected JAK2V617F AB. Conversely, laboratories using a qualitative approach did not detect the positivity of samples with a low AB (0.15%). To further investigate the inter-laboratory variability on low-positive samples, we developed a 2nd round, in which 3 additional laboratories were included. In this 2nd round, each laboratory performed 2 runs with ipsogen JAK2 MutaQuant kit and 2 runs with their routinely used method. None of the laboratories using qualitative “in-house” methods were able to detect low-positive samples, while quantitative results by ipsogen JAK2 MutaQuant kit showed only a small variability among different laboratories at low AB (0.1 and 1%; CV =0.42 and 0.24, respectively). The 3rd round was intended to confirm the robustness of the ipsogen JAK2 MutaQuant kit in a larger cohort of laboratories. The study was therefore extended to 9 additional laboratories. “Home-made” methods were excluded and all laboratories performed 2 runs with the ipsogen JAK2 MutaQuant kit. Quantitative results were well reproducible across all mutation loads. Only one laboratory failed to quantify 0.1% sample in one run. Importantly, all laboratories clearly distinguished between the 0.1 and 1% mutated samples (0.1 and 1%; CV =0.46 and 0.77, respectively).
Conclusion
The first result of the study is that a qualitative approach is not sensitive enough to detect the JAK2V617F mutation at a low (≤1%) burden. Conversely, the ipsogen JAK2 MutaQuant kit resulted highly efficient and sensitive in the quantitative detection of all mutation loads. This study sets the basis for the creation of an Italian network of molecular laboratories focused on the diagnosis of MPNs, including not only JAK2V617F, but also Calreticulin and MPL mutations. The network will aim to identify/standardize the most efficient and cost-effective techniques for the evaluation of these mutations, so to produce reliable and reproducible molecular data.
Session topic: E-poster
Keyword(s): Molecular markers, Myeloproliferative disorder, Real time PCR, Standardization
Type: Publication Only
Background
In chronic myeloproliferative neoplasms (MPNs), the quantification of the JAK2V617F allele burden (AB) is crucial for diagnosis and prognosis assessment, and also for disease monitoring after allogeneic stem-cell transplantation. To-date, a plethora of techniques for JAK2V617F determination is used over different molecular laboratories, with substantial differences in specificity and sensitivity. Given the need to provide reliable and comparable molecular results, the standardization of molecular techniques is of utmost importance.
Aims
The aims of this multicenter study were: 1) to evaluate the inter- and intra-laboratory variability in JAK2V617F quantification in 19 Italian molecular laboratories; 2) to identify the most robust assay for the standardization of the molecular test; 3) to allow consistent interpretation of individual patient analysis results.
Methods
A network of 19 Italian molecular laboratories was established. The study was coordinated by the Institute of Hematology “L. and A. Seràgnoli”, Bologna, and was developed in 3 different rounds (Fig. 1). In routine practice, 2 laboratories did not assess JAK2V617F mutation whereas 7 used a qualitative approach and 10 performed a quantitative evaluation. Both reagents and DNA samples were provided by Werfen-IL SpA and QIAGEN. Raw data and runs validity were checked according to handbook recommendations. Statistical analysis was carried out by QIAGEN/Bologna University.
Results
In the 1st round, we aimed to investigate the inter-laboratory variability on different mutation loads. All laboratories using a quantitative approach were able to determine the expected JAK2V617F AB. Conversely, laboratories using a qualitative approach did not detect the positivity of samples with a low AB (0.15%). To further investigate the inter-laboratory variability on low-positive samples, we developed a 2nd round, in which 3 additional laboratories were included. In this 2nd round, each laboratory performed 2 runs with ipsogen JAK2 MutaQuant kit and 2 runs with their routinely used method. None of the laboratories using qualitative “in-house” methods were able to detect low-positive samples, while quantitative results by ipsogen JAK2 MutaQuant kit showed only a small variability among different laboratories at low AB (0.1 and 1%; CV =0.42 and 0.24, respectively). The 3rd round was intended to confirm the robustness of the ipsogen JAK2 MutaQuant kit in a larger cohort of laboratories. The study was therefore extended to 9 additional laboratories. “Home-made” methods were excluded and all laboratories performed 2 runs with the ipsogen JAK2 MutaQuant kit. Quantitative results were well reproducible across all mutation loads. Only one laboratory failed to quantify 0.1% sample in one run. Importantly, all laboratories clearly distinguished between the 0.1 and 1% mutated samples (0.1 and 1%; CV =0.46 and 0.77, respectively).
Conclusion
The first result of the study is that a qualitative approach is not sensitive enough to detect the JAK2V617F mutation at a low (≤1%) burden. Conversely, the ipsogen JAK2 MutaQuant kit resulted highly efficient and sensitive in the quantitative detection of all mutation loads. This study sets the basis for the creation of an Italian network of molecular laboratories focused on the diagnosis of MPNs, including not only JAK2V617F, but also Calreticulin and MPL mutations. The network will aim to identify/standardize the most efficient and cost-effective techniques for the evaluation of these mutations, so to produce reliable and reproducible molecular data.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloproliferative disorder, Real time PCR, Standardization
Abstract: PB2009
Type: Publication Only
Background
In chronic myeloproliferative neoplasms (MPNs), the quantification of the JAK2V617F allele burden (AB) is crucial for diagnosis and prognosis assessment, and also for disease monitoring after allogeneic stem-cell transplantation. To-date, a plethora of techniques for JAK2V617F determination is used over different molecular laboratories, with substantial differences in specificity and sensitivity. Given the need to provide reliable and comparable molecular results, the standardization of molecular techniques is of utmost importance.
Aims
The aims of this multicenter study were: 1) to evaluate the inter- and intra-laboratory variability in JAK2V617F quantification in 19 Italian molecular laboratories; 2) to identify the most robust assay for the standardization of the molecular test; 3) to allow consistent interpretation of individual patient analysis results.
Methods
A network of 19 Italian molecular laboratories was established. The study was coordinated by the Institute of Hematology “L. and A. Seràgnoli”, Bologna, and was developed in 3 different rounds (Fig. 1). In routine practice, 2 laboratories did not assess JAK2V617F mutation whereas 7 used a qualitative approach and 10 performed a quantitative evaluation. Both reagents and DNA samples were provided by Werfen-IL SpA and QIAGEN. Raw data and runs validity were checked according to handbook recommendations. Statistical analysis was carried out by QIAGEN/Bologna University.
Results
In the 1st round, we aimed to investigate the inter-laboratory variability on different mutation loads. All laboratories using a quantitative approach were able to determine the expected JAK2V617F AB. Conversely, laboratories using a qualitative approach did not detect the positivity of samples with a low AB (0.15%). To further investigate the inter-laboratory variability on low-positive samples, we developed a 2nd round, in which 3 additional laboratories were included. In this 2nd round, each laboratory performed 2 runs with ipsogen JAK2 MutaQuant kit and 2 runs with their routinely used method. None of the laboratories using qualitative “in-house” methods were able to detect low-positive samples, while quantitative results by ipsogen JAK2 MutaQuant kit showed only a small variability among different laboratories at low AB (0.1 and 1%; CV =0.42 and 0.24, respectively). The 3rd round was intended to confirm the robustness of the ipsogen JAK2 MutaQuant kit in a larger cohort of laboratories. The study was therefore extended to 9 additional laboratories. “Home-made” methods were excluded and all laboratories performed 2 runs with the ipsogen JAK2 MutaQuant kit. Quantitative results were well reproducible across all mutation loads. Only one laboratory failed to quantify 0.1% sample in one run. Importantly, all laboratories clearly distinguished between the 0.1 and 1% mutated samples (0.1 and 1%; CV =0.46 and 0.77, respectively).
Conclusion
The first result of the study is that a qualitative approach is not sensitive enough to detect the JAK2V617F mutation at a low (≤1%) burden. Conversely, the ipsogen JAK2 MutaQuant kit resulted highly efficient and sensitive in the quantitative detection of all mutation loads. This study sets the basis for the creation of an Italian network of molecular laboratories focused on the diagnosis of MPNs, including not only JAK2V617F, but also Calreticulin and MPL mutations. The network will aim to identify/standardize the most efficient and cost-effective techniques for the evaluation of these mutations, so to produce reliable and reproducible molecular data.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloproliferative disorder, Real time PCR, Standardization
Type: Publication Only
Background
In chronic myeloproliferative neoplasms (MPNs), the quantification of the JAK2V617F allele burden (AB) is crucial for diagnosis and prognosis assessment, and also for disease monitoring after allogeneic stem-cell transplantation. To-date, a plethora of techniques for JAK2V617F determination is used over different molecular laboratories, with substantial differences in specificity and sensitivity. Given the need to provide reliable and comparable molecular results, the standardization of molecular techniques is of utmost importance.
Aims
The aims of this multicenter study were: 1) to evaluate the inter- and intra-laboratory variability in JAK2V617F quantification in 19 Italian molecular laboratories; 2) to identify the most robust assay for the standardization of the molecular test; 3) to allow consistent interpretation of individual patient analysis results.
Methods
A network of 19 Italian molecular laboratories was established. The study was coordinated by the Institute of Hematology “L. and A. Seràgnoli”, Bologna, and was developed in 3 different rounds (Fig. 1). In routine practice, 2 laboratories did not assess JAK2V617F mutation whereas 7 used a qualitative approach and 10 performed a quantitative evaluation. Both reagents and DNA samples were provided by Werfen-IL SpA and QIAGEN. Raw data and runs validity were checked according to handbook recommendations. Statistical analysis was carried out by QIAGEN/Bologna University.
Results
In the 1st round, we aimed to investigate the inter-laboratory variability on different mutation loads. All laboratories using a quantitative approach were able to determine the expected JAK2V617F AB. Conversely, laboratories using a qualitative approach did not detect the positivity of samples with a low AB (0.15%). To further investigate the inter-laboratory variability on low-positive samples, we developed a 2nd round, in which 3 additional laboratories were included. In this 2nd round, each laboratory performed 2 runs with ipsogen JAK2 MutaQuant kit and 2 runs with their routinely used method. None of the laboratories using qualitative “in-house” methods were able to detect low-positive samples, while quantitative results by ipsogen JAK2 MutaQuant kit showed only a small variability among different laboratories at low AB (0.1 and 1%; CV =0.42 and 0.24, respectively). The 3rd round was intended to confirm the robustness of the ipsogen JAK2 MutaQuant kit in a larger cohort of laboratories. The study was therefore extended to 9 additional laboratories. “Home-made” methods were excluded and all laboratories performed 2 runs with the ipsogen JAK2 MutaQuant kit. Quantitative results were well reproducible across all mutation loads. Only one laboratory failed to quantify 0.1% sample in one run. Importantly, all laboratories clearly distinguished between the 0.1 and 1% mutated samples (0.1 and 1%; CV =0.46 and 0.77, respectively).
Conclusion
The first result of the study is that a qualitative approach is not sensitive enough to detect the JAK2V617F mutation at a low (≤1%) burden. Conversely, the ipsogen JAK2 MutaQuant kit resulted highly efficient and sensitive in the quantitative detection of all mutation loads. This study sets the basis for the creation of an Italian network of molecular laboratories focused on the diagnosis of MPNs, including not only JAK2V617F, but also Calreticulin and MPL mutations. The network will aim to identify/standardize the most efficient and cost-effective techniques for the evaluation of these mutations, so to produce reliable and reproducible molecular data.

Session topic: E-poster
Keyword(s): Molecular markers, Myeloproliferative disorder, Real time PCR, Standardization
{{ help_message }}
{{filter}}


