PLASMA PROTEOMICS IN CHRONIC MYELOID LEUKEMIA PATIENTS BEFORE AND AFTER INITIATION OF TYROSINE KINASE INHIBITOR THERAPY REVEALS INDUCED TH1 IMMUNITY AND LOSS OF ANGIOGENIC STIMULI
(Abstract release date: 05/19/16)
EHA Library. Söderlund S. 06/09/16; 134705; PB1805

Mrs. Stina Söderlund
Contributions
Contributions
Abstract
Abstract: PB1805
Type: Publication Only
Background
Chronic myeloid leukemia (CML) treated with tyrosine kinase inhibitors (TKIs) in most cases have an excellent long-term prognosis. However, there are still problematic issues in a smaller proportion of CML patients related to TKI resistance and long-term treatment side effects and costs. Much is still unknown about why patients respond differently to TKI treatment and why some patients are even able to stop TKI treatment without disease relapse while others relapse quickly despite seemingly good, durable treatment responses.Proteomics is an area of growing interest and the simultaneous measurement of many proteins is now possible using multiplex assays. Multiplexing is used for several purposes, such as surveys of changes in protein abundance, biomarker validation and clinical diagnostics and to our knowledge, the usefulness of plasma proteomics has not been evaluated in CML patients.
Aims
In this pilot study we investigated a total of 124 proteins in plasma from CML patients with the purpose of identifying proteins that are differently expressed at diagnosis and after TKI treatment initiation, either as a result of the decreased disease burden or as an effect of the TKI treatment itself.
Methods
Samples were taken from 14 CML patients at diagnosis and after three months of TKI treatment (imatinib or dasatinib). Samples were analyzed by three different multiplex platforms: Human proinflammatory 9-plex Ultra-Sensitive kit by MesoScale Discovery, Multi-analyte profiling (MAP) technology by Myriad RBM and Proseek Oncology 1 by Olink. Results were correlated to disease activity (Sokal score, presence of Ph+ stem and progenitor cells at diagnosis) and treatment response.
Results
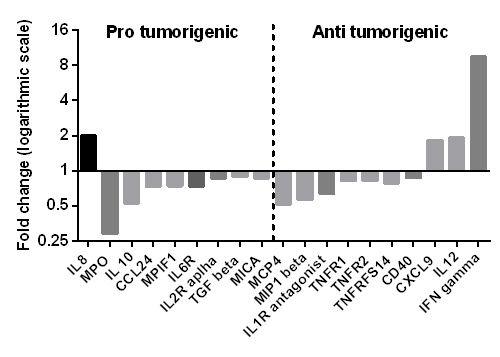
Many protein markers were significantly altered after three months of TKI treatment. Some proteins were analyzed on more than one platform and results for markers present in low concentration in plasma were sometimes contradicting between platforms, which may reflect the specificity and sensitivity variation of the platforms used. It is also highlighting possible difficulties analyzing multiple markers of different concentration ranges in a single sample at one dilution.The protein patterns demonstrated a decrease of pro-tumorigenic analytes (VEGF, TGFβ, IL10, CD31, MICA) while some analytes known to be of importance to T-helper 1 (Th1) immune responses and anti-cancer immunity were increased after TKI initiation (IL12, CXCL9/MIG, IFNγ) (Figure 1), likely reflecting a restoration of normal immune functions after treatment initiation. Interestingly, the level of TGFβ, which has been connected to the maintenance of leukemic stem cells, was correlated to the leukemic stem cell burden (Ph+CD34+CD38- cells) at baseline. We also found reduced angiogenic stimuli which could reflect normalization of bone marrow angiogenesis after treatment initiation since increased bone marrow vascularity and elevated angiogenic factors have been described in untreated CML patients. Further, some single plasma proteins were identified that can be of potential interest to study further for biologic, prognostic or therapeutic significance such as E-selectin, uPAR, growth hormone and carbonic anhydrase IX.
Conclusion
Plasma proteomics seems feasible and useful in CML patients, both for studying patterns of protein expression and for identifying single proteins differentially expressed before and after treatment. Hence, plasma proteomics can be used to gain better understanding of drug mechanisms and treatment responses in CML. Some of the significantly altered proteins indicate novel disease or treatment mechanisms and further studies may give novel insights in CML and TKI therapy.
Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Proteomics, Th1/Th2, Tyrosine kinase inhibitor
Type: Publication Only
Background
Chronic myeloid leukemia (CML) treated with tyrosine kinase inhibitors (TKIs) in most cases have an excellent long-term prognosis. However, there are still problematic issues in a smaller proportion of CML patients related to TKI resistance and long-term treatment side effects and costs. Much is still unknown about why patients respond differently to TKI treatment and why some patients are even able to stop TKI treatment without disease relapse while others relapse quickly despite seemingly good, durable treatment responses.Proteomics is an area of growing interest and the simultaneous measurement of many proteins is now possible using multiplex assays. Multiplexing is used for several purposes, such as surveys of changes in protein abundance, biomarker validation and clinical diagnostics and to our knowledge, the usefulness of plasma proteomics has not been evaluated in CML patients.
Aims
In this pilot study we investigated a total of 124 proteins in plasma from CML patients with the purpose of identifying proteins that are differently expressed at diagnosis and after TKI treatment initiation, either as a result of the decreased disease burden or as an effect of the TKI treatment itself.
Methods
Samples were taken from 14 CML patients at diagnosis and after three months of TKI treatment (imatinib or dasatinib). Samples were analyzed by three different multiplex platforms: Human proinflammatory 9-plex Ultra-Sensitive kit by MesoScale Discovery, Multi-analyte profiling (MAP) technology by Myriad RBM and Proseek Oncology 1 by Olink. Results were correlated to disease activity (Sokal score, presence of Ph+ stem and progenitor cells at diagnosis) and treatment response.
Results
Many protein markers were significantly altered after three months of TKI treatment. Some proteins were analyzed on more than one platform and results for markers present in low concentration in plasma were sometimes contradicting between platforms, which may reflect the specificity and sensitivity variation of the platforms used. It is also highlighting possible difficulties analyzing multiple markers of different concentration ranges in a single sample at one dilution.The protein patterns demonstrated a decrease of pro-tumorigenic analytes (VEGF, TGFβ, IL10, CD31, MICA) while some analytes known to be of importance to T-helper 1 (Th1) immune responses and anti-cancer immunity were increased after TKI initiation (IL12, CXCL9/MIG, IFNγ) (Figure 1), likely reflecting a restoration of normal immune functions after treatment initiation. Interestingly, the level of TGFβ, which has been connected to the maintenance of leukemic stem cells, was correlated to the leukemic stem cell burden (Ph+CD34+CD38- cells) at baseline. We also found reduced angiogenic stimuli which could reflect normalization of bone marrow angiogenesis after treatment initiation since increased bone marrow vascularity and elevated angiogenic factors have been described in untreated CML patients. Further, some single plasma proteins were identified that can be of potential interest to study further for biologic, prognostic or therapeutic significance such as E-selectin, uPAR, growth hormone and carbonic anhydrase IX.
Conclusion
Plasma proteomics seems feasible and useful in CML patients, both for studying patterns of protein expression and for identifying single proteins differentially expressed before and after treatment. Hence, plasma proteomics can be used to gain better understanding of drug mechanisms and treatment responses in CML. Some of the significantly altered proteins indicate novel disease or treatment mechanisms and further studies may give novel insights in CML and TKI therapy.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Proteomics, Th1/Th2, Tyrosine kinase inhibitor
Abstract: PB1805
Type: Publication Only
Background
Chronic myeloid leukemia (CML) treated with tyrosine kinase inhibitors (TKIs) in most cases have an excellent long-term prognosis. However, there are still problematic issues in a smaller proportion of CML patients related to TKI resistance and long-term treatment side effects and costs. Much is still unknown about why patients respond differently to TKI treatment and why some patients are even able to stop TKI treatment without disease relapse while others relapse quickly despite seemingly good, durable treatment responses.Proteomics is an area of growing interest and the simultaneous measurement of many proteins is now possible using multiplex assays. Multiplexing is used for several purposes, such as surveys of changes in protein abundance, biomarker validation and clinical diagnostics and to our knowledge, the usefulness of plasma proteomics has not been evaluated in CML patients.
Aims
In this pilot study we investigated a total of 124 proteins in plasma from CML patients with the purpose of identifying proteins that are differently expressed at diagnosis and after TKI treatment initiation, either as a result of the decreased disease burden or as an effect of the TKI treatment itself.
Methods
Samples were taken from 14 CML patients at diagnosis and after three months of TKI treatment (imatinib or dasatinib). Samples were analyzed by three different multiplex platforms: Human proinflammatory 9-plex Ultra-Sensitive kit by MesoScale Discovery, Multi-analyte profiling (MAP) technology by Myriad RBM and Proseek Oncology 1 by Olink. Results were correlated to disease activity (Sokal score, presence of Ph+ stem and progenitor cells at diagnosis) and treatment response.
Results
Many protein markers were significantly altered after three months of TKI treatment. Some proteins were analyzed on more than one platform and results for markers present in low concentration in plasma were sometimes contradicting between platforms, which may reflect the specificity and sensitivity variation of the platforms used. It is also highlighting possible difficulties analyzing multiple markers of different concentration ranges in a single sample at one dilution.The protein patterns demonstrated a decrease of pro-tumorigenic analytes (VEGF, TGFβ, IL10, CD31, MICA) while some analytes known to be of importance to T-helper 1 (Th1) immune responses and anti-cancer immunity were increased after TKI initiation (IL12, CXCL9/MIG, IFNγ) (Figure 1), likely reflecting a restoration of normal immune functions after treatment initiation. Interestingly, the level of TGFβ, which has been connected to the maintenance of leukemic stem cells, was correlated to the leukemic stem cell burden (Ph+CD34+CD38- cells) at baseline. We also found reduced angiogenic stimuli which could reflect normalization of bone marrow angiogenesis after treatment initiation since increased bone marrow vascularity and elevated angiogenic factors have been described in untreated CML patients. Further, some single plasma proteins were identified that can be of potential interest to study further for biologic, prognostic or therapeutic significance such as E-selectin, uPAR, growth hormone and carbonic anhydrase IX.
Conclusion
Plasma proteomics seems feasible and useful in CML patients, both for studying patterns of protein expression and for identifying single proteins differentially expressed before and after treatment. Hence, plasma proteomics can be used to gain better understanding of drug mechanisms and treatment responses in CML. Some of the significantly altered proteins indicate novel disease or treatment mechanisms and further studies may give novel insights in CML and TKI therapy.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Proteomics, Th1/Th2, Tyrosine kinase inhibitor
Type: Publication Only
Background
Chronic myeloid leukemia (CML) treated with tyrosine kinase inhibitors (TKIs) in most cases have an excellent long-term prognosis. However, there are still problematic issues in a smaller proportion of CML patients related to TKI resistance and long-term treatment side effects and costs. Much is still unknown about why patients respond differently to TKI treatment and why some patients are even able to stop TKI treatment without disease relapse while others relapse quickly despite seemingly good, durable treatment responses.Proteomics is an area of growing interest and the simultaneous measurement of many proteins is now possible using multiplex assays. Multiplexing is used for several purposes, such as surveys of changes in protein abundance, biomarker validation and clinical diagnostics and to our knowledge, the usefulness of plasma proteomics has not been evaluated in CML patients.
Aims
In this pilot study we investigated a total of 124 proteins in plasma from CML patients with the purpose of identifying proteins that are differently expressed at diagnosis and after TKI treatment initiation, either as a result of the decreased disease burden or as an effect of the TKI treatment itself.
Methods
Samples were taken from 14 CML patients at diagnosis and after three months of TKI treatment (imatinib or dasatinib). Samples were analyzed by three different multiplex platforms: Human proinflammatory 9-plex Ultra-Sensitive kit by MesoScale Discovery, Multi-analyte profiling (MAP) technology by Myriad RBM and Proseek Oncology 1 by Olink. Results were correlated to disease activity (Sokal score, presence of Ph+ stem and progenitor cells at diagnosis) and treatment response.
Results
Many protein markers were significantly altered after three months of TKI treatment. Some proteins were analyzed on more than one platform and results for markers present in low concentration in plasma were sometimes contradicting between platforms, which may reflect the specificity and sensitivity variation of the platforms used. It is also highlighting possible difficulties analyzing multiple markers of different concentration ranges in a single sample at one dilution.The protein patterns demonstrated a decrease of pro-tumorigenic analytes (VEGF, TGFβ, IL10, CD31, MICA) while some analytes known to be of importance to T-helper 1 (Th1) immune responses and anti-cancer immunity were increased after TKI initiation (IL12, CXCL9/MIG, IFNγ) (Figure 1), likely reflecting a restoration of normal immune functions after treatment initiation. Interestingly, the level of TGFβ, which has been connected to the maintenance of leukemic stem cells, was correlated to the leukemic stem cell burden (Ph+CD34+CD38- cells) at baseline. We also found reduced angiogenic stimuli which could reflect normalization of bone marrow angiogenesis after treatment initiation since increased bone marrow vascularity and elevated angiogenic factors have been described in untreated CML patients. Further, some single plasma proteins were identified that can be of potential interest to study further for biologic, prognostic or therapeutic significance such as E-selectin, uPAR, growth hormone and carbonic anhydrase IX.
Conclusion
Plasma proteomics seems feasible and useful in CML patients, both for studying patterns of protein expression and for identifying single proteins differentially expressed before and after treatment. Hence, plasma proteomics can be used to gain better understanding of drug mechanisms and treatment responses in CML. Some of the significantly altered proteins indicate novel disease or treatment mechanisms and further studies may give novel insights in CML and TKI therapy.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia, Proteomics, Th1/Th2, Tyrosine kinase inhibitor
{{ help_message }}
{{filter}}


