DYNAMICS OF BCR-ABL1 MULTIPLE MUTATION DETECTED BY SUBCLONING AND SEQUENCING IN TYROSINE KINASE INHIBITOR RESISTANT CML
(Abstract release date: 05/19/16)
EHA Library. Kang K. 06/09/16; 134703; PB1803

Mr. Ki Hoon Kang
Contributions
Contributions
Abstract
Abstract: PB1803
Type: Publication Only
Background
BCR-ABL1 kinase domain (KD) point mutation causes resistance to tyrosine kinase inhibitors (TKI) in CML patients through impaired binding of TKI to the target site. One of the characteristics of patients with BCR-ABL1 kinase domain point mutations is the fact that some patients have multiple mutations. However there have not been many studies showing that data about clinical relevance or dynamics of multiple mutation during CML treatment.
Aims
The aim of this study to was to evaluate dynamics and characteristics of multiple mutations in the serial samples from the patients carrying multiple mutations using subcloning and sequencing.
Methods
Since 2002, 735 CML patients were screened for mutation analysis due to sign of resistance to TKI including imatinib, nilotinib, dasatinib, bosutinib, radotinib or ponatinib at Seoul St Mary’s Hospital using direct sequencing and ASO-PCR. Among them, 42 patients showed multiple mutations. We analyzed serial samples from the 42 patients using subcloning and sequencing to investigate whether the multiple mutations are on same clone (defined as compound mutation), separated clones (defined as polyclonal mutation) and characterize its clinical relevance and dynamics.
Results
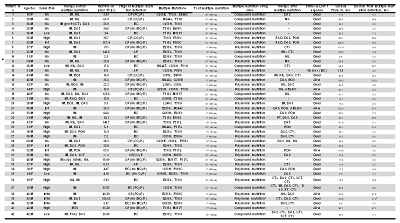
Details on response and survival outcome of 42 patients with multiple mutations is shown in Table 1. In order to investigate whether the multiple mutations are on same clone or on separated clone, we cloned serial samples from the 42 patients. Cloning of cDNA region corresponding to BCR-ABL1 KD into plasmid was performed and followed by transformation into competent cells, colony formation, plasmid preparation of 20 colonies from each sample, and then direct sequencing.Distribution of disease phase at first multiple mutation detection, advanced phase or blast phase was 48% and chronic phase was 52%. With a median follow-up of 35.6 months (range, 4.8-224), 20 different mutations were detected by direct sequencing in 42 patients. 36 patients harbored 2 mutations and 6 patients harbored 3 mutations. Among 42 patients with Multiple mutation, 19(45%) and 23(55%) patients had compound mutation and polyclonal mutation, respectively. of 19 patients with compound mutation, 18 patients(95%) were dead. 26 of 42 patients(62%) developed multiple mutations including T315I. Among them, 16 patients(62%) also had P-loop mutation.
Conclusion
Analysis of serial samples from a same patient provided evidence of dynamic change of portion of compound mutation. In most case, portion of the containing compound mutation was increased as treatment went on, indicating the clone harboring compound mutation can take survival advantage over TKI treatment in comparison of the clone containing individual type of mutation. In addition, some patients showed change in individual mutation type comprising multiple mutations as treatment went on. Patients with compound mutation showed poor outcomes compared with polyclonal mutation in our cohort, further investigation on a large patient cohort will be needed. Updated data about dynamics and characteristic of multiple mutations with longer follow-up duration will be presented in the meeting.* Even though E255k detected by direct sequencing, it was not detected by subcloning and sequencing.
Session topic: E-poster
Keyword(s): Chronic myeloid leukemia
Type: Publication Only
Background
BCR-ABL1 kinase domain (KD) point mutation causes resistance to tyrosine kinase inhibitors (TKI) in CML patients through impaired binding of TKI to the target site. One of the characteristics of patients with BCR-ABL1 kinase domain point mutations is the fact that some patients have multiple mutations. However there have not been many studies showing that data about clinical relevance or dynamics of multiple mutation during CML treatment.
Aims
The aim of this study to was to evaluate dynamics and characteristics of multiple mutations in the serial samples from the patients carrying multiple mutations using subcloning and sequencing.
Methods
Since 2002, 735 CML patients were screened for mutation analysis due to sign of resistance to TKI including imatinib, nilotinib, dasatinib, bosutinib, radotinib or ponatinib at Seoul St Mary’s Hospital using direct sequencing and ASO-PCR. Among them, 42 patients showed multiple mutations. We analyzed serial samples from the 42 patients using subcloning and sequencing to investigate whether the multiple mutations are on same clone (defined as compound mutation), separated clones (defined as polyclonal mutation) and characterize its clinical relevance and dynamics.
Results
Details on response and survival outcome of 42 patients with multiple mutations is shown in Table 1. In order to investigate whether the multiple mutations are on same clone or on separated clone, we cloned serial samples from the 42 patients. Cloning of cDNA region corresponding to BCR-ABL1 KD into plasmid was performed and followed by transformation into competent cells, colony formation, plasmid preparation of 20 colonies from each sample, and then direct sequencing.Distribution of disease phase at first multiple mutation detection, advanced phase or blast phase was 48% and chronic phase was 52%. With a median follow-up of 35.6 months (range, 4.8-224), 20 different mutations were detected by direct sequencing in 42 patients. 36 patients harbored 2 mutations and 6 patients harbored 3 mutations. Among 42 patients with Multiple mutation, 19(45%) and 23(55%) patients had compound mutation and polyclonal mutation, respectively. of 19 patients with compound mutation, 18 patients(95%) were dead. 26 of 42 patients(62%) developed multiple mutations including T315I. Among them, 16 patients(62%) also had P-loop mutation.
Conclusion
Analysis of serial samples from a same patient provided evidence of dynamic change of portion of compound mutation. In most case, portion of the containing compound mutation was increased as treatment went on, indicating the clone harboring compound mutation can take survival advantage over TKI treatment in comparison of the clone containing individual type of mutation. In addition, some patients showed change in individual mutation type comprising multiple mutations as treatment went on. Patients with compound mutation showed poor outcomes compared with polyclonal mutation in our cohort, further investigation on a large patient cohort will be needed. Updated data about dynamics and characteristic of multiple mutations with longer follow-up duration will be presented in the meeting.* Even though E255k detected by direct sequencing, it was not detected by subcloning and sequencing.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia
Abstract: PB1803
Type: Publication Only
Background
BCR-ABL1 kinase domain (KD) point mutation causes resistance to tyrosine kinase inhibitors (TKI) in CML patients through impaired binding of TKI to the target site. One of the characteristics of patients with BCR-ABL1 kinase domain point mutations is the fact that some patients have multiple mutations. However there have not been many studies showing that data about clinical relevance or dynamics of multiple mutation during CML treatment.
Aims
The aim of this study to was to evaluate dynamics and characteristics of multiple mutations in the serial samples from the patients carrying multiple mutations using subcloning and sequencing.
Methods
Since 2002, 735 CML patients were screened for mutation analysis due to sign of resistance to TKI including imatinib, nilotinib, dasatinib, bosutinib, radotinib or ponatinib at Seoul St Mary’s Hospital using direct sequencing and ASO-PCR. Among them, 42 patients showed multiple mutations. We analyzed serial samples from the 42 patients using subcloning and sequencing to investigate whether the multiple mutations are on same clone (defined as compound mutation), separated clones (defined as polyclonal mutation) and characterize its clinical relevance and dynamics.
Results
Details on response and survival outcome of 42 patients with multiple mutations is shown in Table 1. In order to investigate whether the multiple mutations are on same clone or on separated clone, we cloned serial samples from the 42 patients. Cloning of cDNA region corresponding to BCR-ABL1 KD into plasmid was performed and followed by transformation into competent cells, colony formation, plasmid preparation of 20 colonies from each sample, and then direct sequencing.Distribution of disease phase at first multiple mutation detection, advanced phase or blast phase was 48% and chronic phase was 52%. With a median follow-up of 35.6 months (range, 4.8-224), 20 different mutations were detected by direct sequencing in 42 patients. 36 patients harbored 2 mutations and 6 patients harbored 3 mutations. Among 42 patients with Multiple mutation, 19(45%) and 23(55%) patients had compound mutation and polyclonal mutation, respectively. of 19 patients with compound mutation, 18 patients(95%) were dead. 26 of 42 patients(62%) developed multiple mutations including T315I. Among them, 16 patients(62%) also had P-loop mutation.
Conclusion
Analysis of serial samples from a same patient provided evidence of dynamic change of portion of compound mutation. In most case, portion of the containing compound mutation was increased as treatment went on, indicating the clone harboring compound mutation can take survival advantage over TKI treatment in comparison of the clone containing individual type of mutation. In addition, some patients showed change in individual mutation type comprising multiple mutations as treatment went on. Patients with compound mutation showed poor outcomes compared with polyclonal mutation in our cohort, further investigation on a large patient cohort will be needed. Updated data about dynamics and characteristic of multiple mutations with longer follow-up duration will be presented in the meeting.* Even though E255k detected by direct sequencing, it was not detected by subcloning and sequencing.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia
Type: Publication Only
Background
BCR-ABL1 kinase domain (KD) point mutation causes resistance to tyrosine kinase inhibitors (TKI) in CML patients through impaired binding of TKI to the target site. One of the characteristics of patients with BCR-ABL1 kinase domain point mutations is the fact that some patients have multiple mutations. However there have not been many studies showing that data about clinical relevance or dynamics of multiple mutation during CML treatment.
Aims
The aim of this study to was to evaluate dynamics and characteristics of multiple mutations in the serial samples from the patients carrying multiple mutations using subcloning and sequencing.
Methods
Since 2002, 735 CML patients were screened for mutation analysis due to sign of resistance to TKI including imatinib, nilotinib, dasatinib, bosutinib, radotinib or ponatinib at Seoul St Mary’s Hospital using direct sequencing and ASO-PCR. Among them, 42 patients showed multiple mutations. We analyzed serial samples from the 42 patients using subcloning and sequencing to investigate whether the multiple mutations are on same clone (defined as compound mutation), separated clones (defined as polyclonal mutation) and characterize its clinical relevance and dynamics.
Results
Details on response and survival outcome of 42 patients with multiple mutations is shown in Table 1. In order to investigate whether the multiple mutations are on same clone or on separated clone, we cloned serial samples from the 42 patients. Cloning of cDNA region corresponding to BCR-ABL1 KD into plasmid was performed and followed by transformation into competent cells, colony formation, plasmid preparation of 20 colonies from each sample, and then direct sequencing.Distribution of disease phase at first multiple mutation detection, advanced phase or blast phase was 48% and chronic phase was 52%. With a median follow-up of 35.6 months (range, 4.8-224), 20 different mutations were detected by direct sequencing in 42 patients. 36 patients harbored 2 mutations and 6 patients harbored 3 mutations. Among 42 patients with Multiple mutation, 19(45%) and 23(55%) patients had compound mutation and polyclonal mutation, respectively. of 19 patients with compound mutation, 18 patients(95%) were dead. 26 of 42 patients(62%) developed multiple mutations including T315I. Among them, 16 patients(62%) also had P-loop mutation.
Conclusion
Analysis of serial samples from a same patient provided evidence of dynamic change of portion of compound mutation. In most case, portion of the containing compound mutation was increased as treatment went on, indicating the clone harboring compound mutation can take survival advantage over TKI treatment in comparison of the clone containing individual type of mutation. In addition, some patients showed change in individual mutation type comprising multiple mutations as treatment went on. Patients with compound mutation showed poor outcomes compared with polyclonal mutation in our cohort, further investigation on a large patient cohort will be needed. Updated data about dynamics and characteristic of multiple mutations with longer follow-up duration will be presented in the meeting.* Even though E255k detected by direct sequencing, it was not detected by subcloning and sequencing.

Session topic: E-poster
Keyword(s): Chronic myeloid leukemia
{{ help_message }}
{{filter}}


