GENOMIC CHARACTERIZATION OF PATIENTS WITH LOW-RISK ACUTE MYELOID LEUKEMIA (AML)
(Abstract release date: 05/19/16)
EHA Library. Prieto-Conde I. 06/09/16; 134531; PB1631

Ms. Isabel Prieto-Conde
Contributions
Contributions
Abstract
Abstract: PB1631
Type: Publication Only
Background
Not all acute myeloid leukemia (AML) patients classified at diagnosis as cytogenetic and/or molecular favorable risk display a good outcome. In fact, up to 40% of patients with core binding factor (CBF) AML will relapse and not all may be cured. In the same way, a half of NPM1 mutated patients have a relapse during the first 3 years after diagnosis and only biallelic disruption of CEBPA (biCEBPA) is required for a favorable outcome that not avoid a relapse rate of 44%. However, mutations responsible for poor outcome in these previously low-risk classified subsets have not been clearly defined.
Aims
1) To determine the mutational profile of favorable risk AML: CBF-AML, NPM1-mutated AML lacking FLT3-ITD and biCEBPA AML. 2) To identify mutations at diagnosis responsible for poor outcome by comparing patients who experiment relapse vs. patients who do not relapse.
Methods
A cohort of 46 patients with favorable-risk AML (median age 55 years) (PETHEMA AML-99-2010) (Table 1). FLT3-ITD mutations were analyzed by fluorescent PCR and capillary electrophoresis. None of the patients harbored FLT3-ITD. A total of 54 genes were targeted by 568 amplicons that ranged from 225 to 275 bp. The combined coverage was 141 kb in sequence length. Amplicon libraries were prepared by TruSight Myeloid sequencing panel (Illumina, CA) and paired-end sequencing runs were performed on a MiSeq (Illumina) genome sequencer. Minimum depth for reliable analysis was fixed in 100x. Sequences obtained were analyzed with the Variant Studio v2.1 software (Illumina).
Results
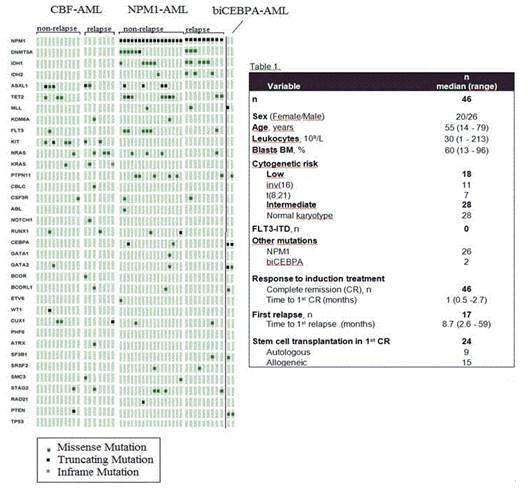
We found 144 mutations in 44 of 46 patients of the global series (3.3 (0-8 mutations/patient) with a mean read depth of 8230x. Only 2 patients remained wild-type for all analyzed genes. Figure 1 displays the mutational distribution of the patients who suffered relapse and patients who do not relapse in the three groups studied. The CBF group (n=18) showed a high frequency of KIT mutations (33%) that were not present in the other low-risk groups. Seven patients harbored NRAS/KRAS mutations (39%), showing a great involvement of RAS pathway in this subset. Other relevant genes affected were ASXL1 and CUX1 (28% and 17 %, respectively). The NPM1 mutated group (n=26) presented a high incidence of DNMT3A, IDH1 and IDH2 mutations, as described previously (35%, 38% and 15%, respectively). Moreover, a 23% of these patients carried mutations in the phosphatase PTPN11 and a 15% in the cohesin complex gene STAG2. The two biCEBPA mutated patients showed altered variants in the transcriptional regulator GATA2. Regarding the prognostic value of these alterations, we found that patients with a better course of the disease had a lower frequency of mutations in NRAS/KRAS genes. Therefore, we analyzed the prognostic value of these variants in the CBF group and it was correlated with a trend to a shorter relapse free survival (3 years, 86% vs. 42% p=0.125). With respect to KIT mutations we did not find significant differences in their clinical adverse impact since 5 of the 6 mutated patients were in first complete remission probably because they underwent stem cell transplantation. Concerning the NPM1 group, mutations were similar in patients with a good outcome and patients who relapse.
Conclusion
This technology is able to find altered variants with high accuracy in AML patients. Although based on small numbers of patients, we observed a clear alteration of the RAS pathway in CBF patients, which suggests the need of further studies about new alternatives to standard chemotherapy in NRAS/KRAS mutated patients.
Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Core binding factor leukemia, Mutation analysis
Type: Publication Only
Background
Not all acute myeloid leukemia (AML) patients classified at diagnosis as cytogenetic and/or molecular favorable risk display a good outcome. In fact, up to 40% of patients with core binding factor (CBF) AML will relapse and not all may be cured. In the same way, a half of NPM1 mutated patients have a relapse during the first 3 years after diagnosis and only biallelic disruption of CEBPA (biCEBPA) is required for a favorable outcome that not avoid a relapse rate of 44%. However, mutations responsible for poor outcome in these previously low-risk classified subsets have not been clearly defined.
Aims
1) To determine the mutational profile of favorable risk AML: CBF-AML, NPM1-mutated AML lacking FLT3-ITD and biCEBPA AML. 2) To identify mutations at diagnosis responsible for poor outcome by comparing patients who experiment relapse vs. patients who do not relapse.
Methods
A cohort of 46 patients with favorable-risk AML (median age 55 years) (PETHEMA AML-99-2010) (Table 1). FLT3-ITD mutations were analyzed by fluorescent PCR and capillary electrophoresis. None of the patients harbored FLT3-ITD. A total of 54 genes were targeted by 568 amplicons that ranged from 225 to 275 bp. The combined coverage was 141 kb in sequence length. Amplicon libraries were prepared by TruSight Myeloid sequencing panel (Illumina, CA) and paired-end sequencing runs were performed on a MiSeq (Illumina) genome sequencer. Minimum depth for reliable analysis was fixed in 100x. Sequences obtained were analyzed with the Variant Studio v2.1 software (Illumina).
Results
We found 144 mutations in 44 of 46 patients of the global series (3.3 (0-8 mutations/patient) with a mean read depth of 8230x. Only 2 patients remained wild-type for all analyzed genes. Figure 1 displays the mutational distribution of the patients who suffered relapse and patients who do not relapse in the three groups studied. The CBF group (n=18) showed a high frequency of KIT mutations (33%) that were not present in the other low-risk groups. Seven patients harbored NRAS/KRAS mutations (39%), showing a great involvement of RAS pathway in this subset. Other relevant genes affected were ASXL1 and CUX1 (28% and 17 %, respectively). The NPM1 mutated group (n=26) presented a high incidence of DNMT3A, IDH1 and IDH2 mutations, as described previously (35%, 38% and 15%, respectively). Moreover, a 23% of these patients carried mutations in the phosphatase PTPN11 and a 15% in the cohesin complex gene STAG2. The two biCEBPA mutated patients showed altered variants in the transcriptional regulator GATA2. Regarding the prognostic value of these alterations, we found that patients with a better course of the disease had a lower frequency of mutations in NRAS/KRAS genes. Therefore, we analyzed the prognostic value of these variants in the CBF group and it was correlated with a trend to a shorter relapse free survival (3 years, 86% vs. 42% p=0.125). With respect to KIT mutations we did not find significant differences in their clinical adverse impact since 5 of the 6 mutated patients were in first complete remission probably because they underwent stem cell transplantation. Concerning the NPM1 group, mutations were similar in patients with a good outcome and patients who relapse.
Conclusion
This technology is able to find altered variants with high accuracy in AML patients. Although based on small numbers of patients, we observed a clear alteration of the RAS pathway in CBF patients, which suggests the need of further studies about new alternatives to standard chemotherapy in NRAS/KRAS mutated patients.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Core binding factor leukemia, Mutation analysis
Abstract: PB1631
Type: Publication Only
Background
Not all acute myeloid leukemia (AML) patients classified at diagnosis as cytogenetic and/or molecular favorable risk display a good outcome. In fact, up to 40% of patients with core binding factor (CBF) AML will relapse and not all may be cured. In the same way, a half of NPM1 mutated patients have a relapse during the first 3 years after diagnosis and only biallelic disruption of CEBPA (biCEBPA) is required for a favorable outcome that not avoid a relapse rate of 44%. However, mutations responsible for poor outcome in these previously low-risk classified subsets have not been clearly defined.
Aims
1) To determine the mutational profile of favorable risk AML: CBF-AML, NPM1-mutated AML lacking FLT3-ITD and biCEBPA AML. 2) To identify mutations at diagnosis responsible for poor outcome by comparing patients who experiment relapse vs. patients who do not relapse.
Methods
A cohort of 46 patients with favorable-risk AML (median age 55 years) (PETHEMA AML-99-2010) (Table 1). FLT3-ITD mutations were analyzed by fluorescent PCR and capillary electrophoresis. None of the patients harbored FLT3-ITD. A total of 54 genes were targeted by 568 amplicons that ranged from 225 to 275 bp. The combined coverage was 141 kb in sequence length. Amplicon libraries were prepared by TruSight Myeloid sequencing panel (Illumina, CA) and paired-end sequencing runs were performed on a MiSeq (Illumina) genome sequencer. Minimum depth for reliable analysis was fixed in 100x. Sequences obtained were analyzed with the Variant Studio v2.1 software (Illumina).
Results
We found 144 mutations in 44 of 46 patients of the global series (3.3 (0-8 mutations/patient) with a mean read depth of 8230x. Only 2 patients remained wild-type for all analyzed genes. Figure 1 displays the mutational distribution of the patients who suffered relapse and patients who do not relapse in the three groups studied. The CBF group (n=18) showed a high frequency of KIT mutations (33%) that were not present in the other low-risk groups. Seven patients harbored NRAS/KRAS mutations (39%), showing a great involvement of RAS pathway in this subset. Other relevant genes affected were ASXL1 and CUX1 (28% and 17 %, respectively). The NPM1 mutated group (n=26) presented a high incidence of DNMT3A, IDH1 and IDH2 mutations, as described previously (35%, 38% and 15%, respectively). Moreover, a 23% of these patients carried mutations in the phosphatase PTPN11 and a 15% in the cohesin complex gene STAG2. The two biCEBPA mutated patients showed altered variants in the transcriptional regulator GATA2. Regarding the prognostic value of these alterations, we found that patients with a better course of the disease had a lower frequency of mutations in NRAS/KRAS genes. Therefore, we analyzed the prognostic value of these variants in the CBF group and it was correlated with a trend to a shorter relapse free survival (3 years, 86% vs. 42% p=0.125). With respect to KIT mutations we did not find significant differences in their clinical adverse impact since 5 of the 6 mutated patients were in first complete remission probably because they underwent stem cell transplantation. Concerning the NPM1 group, mutations were similar in patients with a good outcome and patients who relapse.
Conclusion
This technology is able to find altered variants with high accuracy in AML patients. Although based on small numbers of patients, we observed a clear alteration of the RAS pathway in CBF patients, which suggests the need of further studies about new alternatives to standard chemotherapy in NRAS/KRAS mutated patients.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Core binding factor leukemia, Mutation analysis
Type: Publication Only
Background
Not all acute myeloid leukemia (AML) patients classified at diagnosis as cytogenetic and/or molecular favorable risk display a good outcome. In fact, up to 40% of patients with core binding factor (CBF) AML will relapse and not all may be cured. In the same way, a half of NPM1 mutated patients have a relapse during the first 3 years after diagnosis and only biallelic disruption of CEBPA (biCEBPA) is required for a favorable outcome that not avoid a relapse rate of 44%. However, mutations responsible for poor outcome in these previously low-risk classified subsets have not been clearly defined.
Aims
1) To determine the mutational profile of favorable risk AML: CBF-AML, NPM1-mutated AML lacking FLT3-ITD and biCEBPA AML. 2) To identify mutations at diagnosis responsible for poor outcome by comparing patients who experiment relapse vs. patients who do not relapse.
Methods
A cohort of 46 patients with favorable-risk AML (median age 55 years) (PETHEMA AML-99-2010) (Table 1). FLT3-ITD mutations were analyzed by fluorescent PCR and capillary electrophoresis. None of the patients harbored FLT3-ITD. A total of 54 genes were targeted by 568 amplicons that ranged from 225 to 275 bp. The combined coverage was 141 kb in sequence length. Amplicon libraries were prepared by TruSight Myeloid sequencing panel (Illumina, CA) and paired-end sequencing runs were performed on a MiSeq (Illumina) genome sequencer. Minimum depth for reliable analysis was fixed in 100x. Sequences obtained were analyzed with the Variant Studio v2.1 software (Illumina).
Results
We found 144 mutations in 44 of 46 patients of the global series (3.3 (0-8 mutations/patient) with a mean read depth of 8230x. Only 2 patients remained wild-type for all analyzed genes. Figure 1 displays the mutational distribution of the patients who suffered relapse and patients who do not relapse in the three groups studied. The CBF group (n=18) showed a high frequency of KIT mutations (33%) that were not present in the other low-risk groups. Seven patients harbored NRAS/KRAS mutations (39%), showing a great involvement of RAS pathway in this subset. Other relevant genes affected were ASXL1 and CUX1 (28% and 17 %, respectively). The NPM1 mutated group (n=26) presented a high incidence of DNMT3A, IDH1 and IDH2 mutations, as described previously (35%, 38% and 15%, respectively). Moreover, a 23% of these patients carried mutations in the phosphatase PTPN11 and a 15% in the cohesin complex gene STAG2. The two biCEBPA mutated patients showed altered variants in the transcriptional regulator GATA2. Regarding the prognostic value of these alterations, we found that patients with a better course of the disease had a lower frequency of mutations in NRAS/KRAS genes. Therefore, we analyzed the prognostic value of these variants in the CBF group and it was correlated with a trend to a shorter relapse free survival (3 years, 86% vs. 42% p=0.125). With respect to KIT mutations we did not find significant differences in their clinical adverse impact since 5 of the 6 mutated patients were in first complete remission probably because they underwent stem cell transplantation. Concerning the NPM1 group, mutations were similar in patients with a good outcome and patients who relapse.
Conclusion
This technology is able to find altered variants with high accuracy in AML patients. Although based on small numbers of patients, we observed a clear alteration of the RAS pathway in CBF patients, which suggests the need of further studies about new alternatives to standard chemotherapy in NRAS/KRAS mutated patients.

Session topic: E-poster
Keyword(s): Acute myeloid leukemia, Core binding factor leukemia, Mutation analysis
{{ help_message }}
{{filter}}


