COMPARATIVE ANALYSIS OF CLONAL IG AND TCR GENE REARRANGEMENTS IN ADULT PH-NEGATIVE ALL AT THE DIAGNOSIS AND AT THE RELAPSE
(Abstract release date: 05/19/16)
EHA Library. Sudarikov A. 06/09/16; 134504; PB1604

Dr. Andrey Sudarikov
Contributions
Contributions
Abstract
Abstract: PB1604
Type: Publication Only
Background
Clonal rearrangements of immunoglobulin (IG) and T-cell receptor (TCR) genes are the targets for minimal residual disease (MRD) assessment. The level of the MRD is an independent prognostic factor and its monitoring is important to identify patients with high risk of relapse. One of the pitfalls of MRD detection by means of IG and TCR gene rearrangements in ALL is clonal evolution possibly caused by V(D)J recombinase activity. Several studies have showed that the pattern of IG and TCR gene rearrangements in ALL patients may change during the course of disease. In relapse some gene rearrangements may be lost while new gene rearrangements may occur. Change of clonal rearrangements, during tumor progression may lead to the loss of a target for MRD studies and false-negative results.
Aims
Studying the phenomenon of clonal heterogeneity in adult patients with ALL at the diagnosis and relapse of disease.
Methods
We have determined T- and B-cell clonality in six adults with Ph-negative ALL at the time of diagnosis and at the relapse. B-cell clonality was assessed by immunoglobulin heavy chain gene rearrangements IGH (VH – JH) and light kappa chain gene rearrangements IGK (Vk – Jk / Vk – KDE / Intron RSS – KDE). T-cell clonality was assessed by T-cell receptor gamma chain TCRG (VG – JG), beta chain TCRB (VB – JB /DB – JB) and delta chain TCRD (VD – JD / DD2 – JD/ VD – DD3/ DD2 – DD3) gene rearrangements. Multiplex BIOMED-2 primer system for PCR and Gene-Scan analysis have been used.
Results
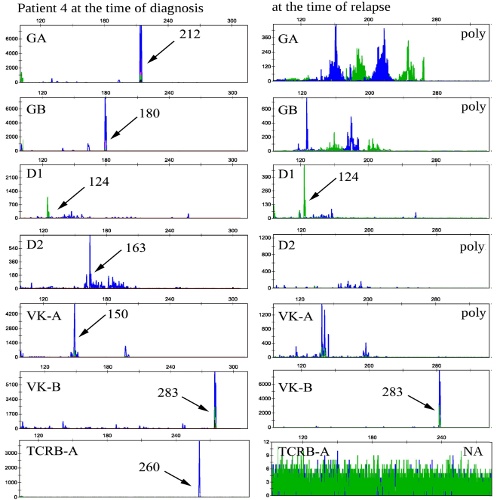
In two patients with B-cell ALL one of clonal rearrangement presented at the diagnosis was lost at the relapse of disease while new rearrangements emerged. In one patient with a diagnosis of early T-ALL clonal rearrangements of the genes TCRD, TCRB and TCRG, identified at the diagnosis were exactly the same in relapse. In one patient all the rearrangements identified at the diagnosis were preserved at the relapse and several new rearrangements appeared. In one case five rearrangements disappeared at the relapse of disease out of seven identified at the diagnosis (fig.1). In one patient only one clonal rearrangement was revealed at the diagnosis of B-cell ALL and at the relapse of the disease this rearrangement was preserved, but there were a few new. To analyze initial ALL samples for possible presence of minor clones that lately we have found in relapse we used appropriate family-specific primers to V and J-region instead of multiplex primer mixtures. Despite increased to 10-2-10-3 sensitivity no detectable cell populations of these subclones were found.
Conclusion
Mismatch in clonal rearrangements at the diagnosis and at the relapse of disease was identified in five of six (83%) patients, indicating clonal instability during the course of treatment. Clonal evolution and diversity of clonal IG and TCR gene rearrangements could be one of the features of tumor progression. Also it could be speculated that many clonal IG and TCR gene rearrangements may present at different amounts at the diagnosis, however less abundant clones could be “invisible” due to the limitation of measuring sensitivity. Later major clones may disappear under the influence of chemotherapy while others could gain proliferation capacity. Study of the clonal evolution and heterogeneity in ALL and their impact on treatment efficacy should help to develop new prognostic factors and therapeutic strategies.
Session topic: E-poster
Keyword(s): ALL, Clonality, PCR, Relapsed acute lymphoblastic leukemia
Type: Publication Only
Background
Clonal rearrangements of immunoglobulin (IG) and T-cell receptor (TCR) genes are the targets for minimal residual disease (MRD) assessment. The level of the MRD is an independent prognostic factor and its monitoring is important to identify patients with high risk of relapse. One of the pitfalls of MRD detection by means of IG and TCR gene rearrangements in ALL is clonal evolution possibly caused by V(D)J recombinase activity. Several studies have showed that the pattern of IG and TCR gene rearrangements in ALL patients may change during the course of disease. In relapse some gene rearrangements may be lost while new gene rearrangements may occur. Change of clonal rearrangements, during tumor progression may lead to the loss of a target for MRD studies and false-negative results.
Aims
Studying the phenomenon of clonal heterogeneity in adult patients with ALL at the diagnosis and relapse of disease.
Methods
We have determined T- and B-cell clonality in six adults with Ph-negative ALL at the time of diagnosis and at the relapse. B-cell clonality was assessed by immunoglobulin heavy chain gene rearrangements IGH (VH – JH) and light kappa chain gene rearrangements IGK (Vk – Jk / Vk – KDE / Intron RSS – KDE). T-cell clonality was assessed by T-cell receptor gamma chain TCRG (VG – JG), beta chain TCRB (VB – JB /DB – JB) and delta chain TCRD (VD – JD / DD2 – JD/ VD – DD3/ DD2 – DD3) gene rearrangements. Multiplex BIOMED-2 primer system for PCR and Gene-Scan analysis have been used.
Results
In two patients with B-cell ALL one of clonal rearrangement presented at the diagnosis was lost at the relapse of disease while new rearrangements emerged. In one patient with a diagnosis of early T-ALL clonal rearrangements of the genes TCRD, TCRB and TCRG, identified at the diagnosis were exactly the same in relapse. In one patient all the rearrangements identified at the diagnosis were preserved at the relapse and several new rearrangements appeared. In one case five rearrangements disappeared at the relapse of disease out of seven identified at the diagnosis (fig.1). In one patient only one clonal rearrangement was revealed at the diagnosis of B-cell ALL and at the relapse of the disease this rearrangement was preserved, but there were a few new. To analyze initial ALL samples for possible presence of minor clones that lately we have found in relapse we used appropriate family-specific primers to V and J-region instead of multiplex primer mixtures. Despite increased to 10-2-10-3 sensitivity no detectable cell populations of these subclones were found.
Conclusion
Mismatch in clonal rearrangements at the diagnosis and at the relapse of disease was identified in five of six (83%) patients, indicating clonal instability during the course of treatment. Clonal evolution and diversity of clonal IG and TCR gene rearrangements could be one of the features of tumor progression. Also it could be speculated that many clonal IG and TCR gene rearrangements may present at different amounts at the diagnosis, however less abundant clones could be “invisible” due to the limitation of measuring sensitivity. Later major clones may disappear under the influence of chemotherapy while others could gain proliferation capacity. Study of the clonal evolution and heterogeneity in ALL and their impact on treatment efficacy should help to develop new prognostic factors and therapeutic strategies.

Session topic: E-poster
Keyword(s): ALL, Clonality, PCR, Relapsed acute lymphoblastic leukemia
Abstract: PB1604
Type: Publication Only
Background
Clonal rearrangements of immunoglobulin (IG) and T-cell receptor (TCR) genes are the targets for minimal residual disease (MRD) assessment. The level of the MRD is an independent prognostic factor and its monitoring is important to identify patients with high risk of relapse. One of the pitfalls of MRD detection by means of IG and TCR gene rearrangements in ALL is clonal evolution possibly caused by V(D)J recombinase activity. Several studies have showed that the pattern of IG and TCR gene rearrangements in ALL patients may change during the course of disease. In relapse some gene rearrangements may be lost while new gene rearrangements may occur. Change of clonal rearrangements, during tumor progression may lead to the loss of a target for MRD studies and false-negative results.
Aims
Studying the phenomenon of clonal heterogeneity in adult patients with ALL at the diagnosis and relapse of disease.
Methods
We have determined T- and B-cell clonality in six adults with Ph-negative ALL at the time of diagnosis and at the relapse. B-cell clonality was assessed by immunoglobulin heavy chain gene rearrangements IGH (VH – JH) and light kappa chain gene rearrangements IGK (Vk – Jk / Vk – KDE / Intron RSS – KDE). T-cell clonality was assessed by T-cell receptor gamma chain TCRG (VG – JG), beta chain TCRB (VB – JB /DB – JB) and delta chain TCRD (VD – JD / DD2 – JD/ VD – DD3/ DD2 – DD3) gene rearrangements. Multiplex BIOMED-2 primer system for PCR and Gene-Scan analysis have been used.
Results
In two patients with B-cell ALL one of clonal rearrangement presented at the diagnosis was lost at the relapse of disease while new rearrangements emerged. In one patient with a diagnosis of early T-ALL clonal rearrangements of the genes TCRD, TCRB and TCRG, identified at the diagnosis were exactly the same in relapse. In one patient all the rearrangements identified at the diagnosis were preserved at the relapse and several new rearrangements appeared. In one case five rearrangements disappeared at the relapse of disease out of seven identified at the diagnosis (fig.1). In one patient only one clonal rearrangement was revealed at the diagnosis of B-cell ALL and at the relapse of the disease this rearrangement was preserved, but there were a few new. To analyze initial ALL samples for possible presence of minor clones that lately we have found in relapse we used appropriate family-specific primers to V and J-region instead of multiplex primer mixtures. Despite increased to 10-2-10-3 sensitivity no detectable cell populations of these subclones were found.
Conclusion
Mismatch in clonal rearrangements at the diagnosis and at the relapse of disease was identified in five of six (83%) patients, indicating clonal instability during the course of treatment. Clonal evolution and diversity of clonal IG and TCR gene rearrangements could be one of the features of tumor progression. Also it could be speculated that many clonal IG and TCR gene rearrangements may present at different amounts at the diagnosis, however less abundant clones could be “invisible” due to the limitation of measuring sensitivity. Later major clones may disappear under the influence of chemotherapy while others could gain proliferation capacity. Study of the clonal evolution and heterogeneity in ALL and their impact on treatment efficacy should help to develop new prognostic factors and therapeutic strategies.

Session topic: E-poster
Keyword(s): ALL, Clonality, PCR, Relapsed acute lymphoblastic leukemia
Type: Publication Only
Background
Clonal rearrangements of immunoglobulin (IG) and T-cell receptor (TCR) genes are the targets for minimal residual disease (MRD) assessment. The level of the MRD is an independent prognostic factor and its monitoring is important to identify patients with high risk of relapse. One of the pitfalls of MRD detection by means of IG and TCR gene rearrangements in ALL is clonal evolution possibly caused by V(D)J recombinase activity. Several studies have showed that the pattern of IG and TCR gene rearrangements in ALL patients may change during the course of disease. In relapse some gene rearrangements may be lost while new gene rearrangements may occur. Change of clonal rearrangements, during tumor progression may lead to the loss of a target for MRD studies and false-negative results.
Aims
Studying the phenomenon of clonal heterogeneity in adult patients with ALL at the diagnosis and relapse of disease.
Methods
We have determined T- and B-cell clonality in six adults with Ph-negative ALL at the time of diagnosis and at the relapse. B-cell clonality was assessed by immunoglobulin heavy chain gene rearrangements IGH (VH – JH) and light kappa chain gene rearrangements IGK (Vk – Jk / Vk – KDE / Intron RSS – KDE). T-cell clonality was assessed by T-cell receptor gamma chain TCRG (VG – JG), beta chain TCRB (VB – JB /DB – JB) and delta chain TCRD (VD – JD / DD2 – JD/ VD – DD3/ DD2 – DD3) gene rearrangements. Multiplex BIOMED-2 primer system for PCR and Gene-Scan analysis have been used.
Results
In two patients with B-cell ALL one of clonal rearrangement presented at the diagnosis was lost at the relapse of disease while new rearrangements emerged. In one patient with a diagnosis of early T-ALL clonal rearrangements of the genes TCRD, TCRB and TCRG, identified at the diagnosis were exactly the same in relapse. In one patient all the rearrangements identified at the diagnosis were preserved at the relapse and several new rearrangements appeared. In one case five rearrangements disappeared at the relapse of disease out of seven identified at the diagnosis (fig.1). In one patient only one clonal rearrangement was revealed at the diagnosis of B-cell ALL and at the relapse of the disease this rearrangement was preserved, but there were a few new. To analyze initial ALL samples for possible presence of minor clones that lately we have found in relapse we used appropriate family-specific primers to V and J-region instead of multiplex primer mixtures. Despite increased to 10-2-10-3 sensitivity no detectable cell populations of these subclones were found.
Conclusion
Mismatch in clonal rearrangements at the diagnosis and at the relapse of disease was identified in five of six (83%) patients, indicating clonal instability during the course of treatment. Clonal evolution and diversity of clonal IG and TCR gene rearrangements could be one of the features of tumor progression. Also it could be speculated that many clonal IG and TCR gene rearrangements may present at different amounts at the diagnosis, however less abundant clones could be “invisible” due to the limitation of measuring sensitivity. Later major clones may disappear under the influence of chemotherapy while others could gain proliferation capacity. Study of the clonal evolution and heterogeneity in ALL and their impact on treatment efficacy should help to develop new prognostic factors and therapeutic strategies.

Session topic: E-poster
Keyword(s): ALL, Clonality, PCR, Relapsed acute lymphoblastic leukemia
{{ help_message }}
{{filter}}


