WHOLE EXOME SEQUENCING (WES) IN PHILADELPHIA NEGATIVE (PH-) ACUTE LYMPHOBLASTIC LEUKEMIA (ALL) EXTRAMEDULLARY RELAPSES IDENTIFIED COMMON JAK2 MUTATIONS
(Abstract release date: 05/19/16)
EHA Library. Ferrari A. 06/09/16; 134495; PB1595

Anna Ferrari
Contributions
Contributions
Abstract
Abstract: PB1595
Type: Publication Only
Background
Acute lymphoblastic leukemia (ALL) is a complex disease with multiple factors related both to the disease itself and host biology that significantly affect outcome. Appropriate risk stratification and a better understanding of the genomic landscape underlying this disease have led to improvement in overall survival in children. Even with these advances, outcome remains unsatisfactory in adults, mainly in the Philadelphia chromosome negative subgroup. Here we report a 22-year-old man with pre-B ALL who was negative for the recurrent known molecular rearrangements (E2A-PBX, TEL, AML1- MLL-AF4). The patient (pt) received different therapeutic regimens and allogenic stem cell transplantation but he did not achieve a remission. Fourteen months after diagnosis (dx), leukemia extramedullary relapsed at the breast and later at lymph node in the groin (CD34+, TdT+,PAX5+, CD19+, CD22+, CD3-).
Aims
to dissect the genetic landscape in order to predict treatment failure and identify targets amenable to inhibition by targeted therapies we compared the genomic profile of 3 different tissues.
Methods
Whole exome sequencing (WES) was performed on dx bone marrow (BM), breast (BR) and lymph node (LN) samples at the time of relapse using the Illumina Hiseq2000 platform. Matched samples of primary tumour and germline DNA from buccal swab were also analyzed. The post-transplantation BR and LN samples were further matched with donor peripheral blood DNA. MuTect and Varscan tools to call mutations (Single Nucleotide Variants=SNVs and/or INDELs) were used. 3 sample SNP array analysis were also performed.
Results
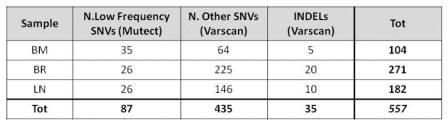
WES analysis from 3 samples identified 522 point mutations and 35 indels that occur in 474 genes (Table 1). The number of total mutations at the time of relapse is higher both in BR and LN samples compare to dx BM. The majority of mutations occurred on genes localized on chr1 (74); the mean number of mutations/chr is 24 (2-74). BR and LN sample have 57 mutations in common (5 in BM and BR, 2 in BM and LN). Mutect identified 26 mutations in both relapsed samples. They include a mutation in FAAH gene, which is also found fused with NSUN4 at the dx (RNASeq). This analysis revealed some other fused genes that were detected mutated with WES. Nine genes are confirmed to be mutated in all 3 samples: 7 nonsynonymous SNVs (ACHE, COL4A2, LOC554223, NPHS1, SLC36A1, TMEM89 and JAK2), 1 stopgain mutation (NTNG2) and 1 indel (HELZ2). Excluding JAK2, no direct relations between these genes and leukemia were reported so far. The JAK2 mutation S1032Y were detected in all 3 samples, instead the R683G one in the BM and BR. CD8 is and presents 2 mutations in both BM and BR. SNP array analysis also reveals that this gene was deleted in heterozygosity in the BM and LN. One copy of EBF1, INPP4B, ZCCHC7 genes are deleted in all 3 samples (SNP array). EBF1 is described to be associated to ALL. So far, we confirmed ZCCHC7 is fused with PAX5 in BM and BR, as previously described (Roberts KG, 2012).
Conclusion
The Ph- ALL extramedullary relapses show higher genetic instability, with similar low frequency mutation profile. WES suggest that JAK2 alterations are driver mutations. This finding could be helpful for a targeted therapy. Supported by: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.
Session topic: E-poster
Keyword(s): Acute lymphoblastic leukemia
Type: Publication Only
Background
Acute lymphoblastic leukemia (ALL) is a complex disease with multiple factors related both to the disease itself and host biology that significantly affect outcome. Appropriate risk stratification and a better understanding of the genomic landscape underlying this disease have led to improvement in overall survival in children. Even with these advances, outcome remains unsatisfactory in adults, mainly in the Philadelphia chromosome negative subgroup. Here we report a 22-year-old man with pre-B ALL who was negative for the recurrent known molecular rearrangements (E2A-PBX, TEL, AML1- MLL-AF4). The patient (pt) received different therapeutic regimens and allogenic stem cell transplantation but he did not achieve a remission. Fourteen months after diagnosis (dx), leukemia extramedullary relapsed at the breast and later at lymph node in the groin (CD34+, TdT+,PAX5+, CD19+, CD22+, CD3-).
Aims
to dissect the genetic landscape in order to predict treatment failure and identify targets amenable to inhibition by targeted therapies we compared the genomic profile of 3 different tissues.
Methods
Whole exome sequencing (WES) was performed on dx bone marrow (BM), breast (BR) and lymph node (LN) samples at the time of relapse using the Illumina Hiseq2000 platform. Matched samples of primary tumour and germline DNA from buccal swab were also analyzed. The post-transplantation BR and LN samples were further matched with donor peripheral blood DNA. MuTect and Varscan tools to call mutations (Single Nucleotide Variants=SNVs and/or INDELs) were used. 3 sample SNP array analysis were also performed.
Results
WES analysis from 3 samples identified 522 point mutations and 35 indels that occur in 474 genes (Table 1). The number of total mutations at the time of relapse is higher both in BR and LN samples compare to dx BM. The majority of mutations occurred on genes localized on chr1 (74); the mean number of mutations/chr is 24 (2-74). BR and LN sample have 57 mutations in common (5 in BM and BR, 2 in BM and LN). Mutect identified 26 mutations in both relapsed samples. They include a mutation in FAAH gene, which is also found fused with NSUN4 at the dx (RNASeq). This analysis revealed some other fused genes that were detected mutated with WES. Nine genes are confirmed to be mutated in all 3 samples: 7 nonsynonymous SNVs (ACHE, COL4A2, LOC554223, NPHS1, SLC36A1, TMEM89 and JAK2), 1 stopgain mutation (NTNG2) and 1 indel (HELZ2). Excluding JAK2, no direct relations between these genes and leukemia were reported so far. The JAK2 mutation S1032Y were detected in all 3 samples, instead the R683G one in the BM and BR. CD8 is and presents 2 mutations in both BM and BR. SNP array analysis also reveals that this gene was deleted in heterozygosity in the BM and LN. One copy of EBF1, INPP4B, ZCCHC7 genes are deleted in all 3 samples (SNP array). EBF1 is described to be associated to ALL. So far, we confirmed ZCCHC7 is fused with PAX5 in BM and BR, as previously described (Roberts KG, 2012).
Conclusion
The Ph- ALL extramedullary relapses show higher genetic instability, with similar low frequency mutation profile. WES suggest that JAK2 alterations are driver mutations. This finding could be helpful for a targeted therapy. Supported by: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute lymphoblastic leukemia
Abstract: PB1595
Type: Publication Only
Background
Acute lymphoblastic leukemia (ALL) is a complex disease with multiple factors related both to the disease itself and host biology that significantly affect outcome. Appropriate risk stratification and a better understanding of the genomic landscape underlying this disease have led to improvement in overall survival in children. Even with these advances, outcome remains unsatisfactory in adults, mainly in the Philadelphia chromosome negative subgroup. Here we report a 22-year-old man with pre-B ALL who was negative for the recurrent known molecular rearrangements (E2A-PBX, TEL, AML1- MLL-AF4). The patient (pt) received different therapeutic regimens and allogenic stem cell transplantation but he did not achieve a remission. Fourteen months after diagnosis (dx), leukemia extramedullary relapsed at the breast and later at lymph node in the groin (CD34+, TdT+,PAX5+, CD19+, CD22+, CD3-).
Aims
to dissect the genetic landscape in order to predict treatment failure and identify targets amenable to inhibition by targeted therapies we compared the genomic profile of 3 different tissues.
Methods
Whole exome sequencing (WES) was performed on dx bone marrow (BM), breast (BR) and lymph node (LN) samples at the time of relapse using the Illumina Hiseq2000 platform. Matched samples of primary tumour and germline DNA from buccal swab were also analyzed. The post-transplantation BR and LN samples were further matched with donor peripheral blood DNA. MuTect and Varscan tools to call mutations (Single Nucleotide Variants=SNVs and/or INDELs) were used. 3 sample SNP array analysis were also performed.
Results
WES analysis from 3 samples identified 522 point mutations and 35 indels that occur in 474 genes (Table 1). The number of total mutations at the time of relapse is higher both in BR and LN samples compare to dx BM. The majority of mutations occurred on genes localized on chr1 (74); the mean number of mutations/chr is 24 (2-74). BR and LN sample have 57 mutations in common (5 in BM and BR, 2 in BM and LN). Mutect identified 26 mutations in both relapsed samples. They include a mutation in FAAH gene, which is also found fused with NSUN4 at the dx (RNASeq). This analysis revealed some other fused genes that were detected mutated with WES. Nine genes are confirmed to be mutated in all 3 samples: 7 nonsynonymous SNVs (ACHE, COL4A2, LOC554223, NPHS1, SLC36A1, TMEM89 and JAK2), 1 stopgain mutation (NTNG2) and 1 indel (HELZ2). Excluding JAK2, no direct relations between these genes and leukemia were reported so far. The JAK2 mutation S1032Y were detected in all 3 samples, instead the R683G one in the BM and BR. CD8 is and presents 2 mutations in both BM and BR. SNP array analysis also reveals that this gene was deleted in heterozygosity in the BM and LN. One copy of EBF1, INPP4B, ZCCHC7 genes are deleted in all 3 samples (SNP array). EBF1 is described to be associated to ALL. So far, we confirmed ZCCHC7 is fused with PAX5 in BM and BR, as previously described (Roberts KG, 2012).
Conclusion
The Ph- ALL extramedullary relapses show higher genetic instability, with similar low frequency mutation profile. WES suggest that JAK2 alterations are driver mutations. This finding could be helpful for a targeted therapy. Supported by: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute lymphoblastic leukemia
Type: Publication Only
Background
Acute lymphoblastic leukemia (ALL) is a complex disease with multiple factors related both to the disease itself and host biology that significantly affect outcome. Appropriate risk stratification and a better understanding of the genomic landscape underlying this disease have led to improvement in overall survival in children. Even with these advances, outcome remains unsatisfactory in adults, mainly in the Philadelphia chromosome negative subgroup. Here we report a 22-year-old man with pre-B ALL who was negative for the recurrent known molecular rearrangements (E2A-PBX, TEL, AML1- MLL-AF4). The patient (pt) received different therapeutic regimens and allogenic stem cell transplantation but he did not achieve a remission. Fourteen months after diagnosis (dx), leukemia extramedullary relapsed at the breast and later at lymph node in the groin (CD34+, TdT+,PAX5+, CD19+, CD22+, CD3-).
Aims
to dissect the genetic landscape in order to predict treatment failure and identify targets amenable to inhibition by targeted therapies we compared the genomic profile of 3 different tissues.
Methods
Whole exome sequencing (WES) was performed on dx bone marrow (BM), breast (BR) and lymph node (LN) samples at the time of relapse using the Illumina Hiseq2000 platform. Matched samples of primary tumour and germline DNA from buccal swab were also analyzed. The post-transplantation BR and LN samples were further matched with donor peripheral blood DNA. MuTect and Varscan tools to call mutations (Single Nucleotide Variants=SNVs and/or INDELs) were used. 3 sample SNP array analysis were also performed.
Results
WES analysis from 3 samples identified 522 point mutations and 35 indels that occur in 474 genes (Table 1). The number of total mutations at the time of relapse is higher both in BR and LN samples compare to dx BM. The majority of mutations occurred on genes localized on chr1 (74); the mean number of mutations/chr is 24 (2-74). BR and LN sample have 57 mutations in common (5 in BM and BR, 2 in BM and LN). Mutect identified 26 mutations in both relapsed samples. They include a mutation in FAAH gene, which is also found fused with NSUN4 at the dx (RNASeq). This analysis revealed some other fused genes that were detected mutated with WES. Nine genes are confirmed to be mutated in all 3 samples: 7 nonsynonymous SNVs (ACHE, COL4A2, LOC554223, NPHS1, SLC36A1, TMEM89 and JAK2), 1 stopgain mutation (NTNG2) and 1 indel (HELZ2). Excluding JAK2, no direct relations between these genes and leukemia were reported so far. The JAK2 mutation S1032Y were detected in all 3 samples, instead the R683G one in the BM and BR. CD8 is and presents 2 mutations in both BM and BR. SNP array analysis also reveals that this gene was deleted in heterozygosity in the BM and LN. One copy of EBF1, INPP4B, ZCCHC7 genes are deleted in all 3 samples (SNP array). EBF1 is described to be associated to ALL. So far, we confirmed ZCCHC7 is fused with PAX5 in BM and BR, as previously described (Roberts KG, 2012).
Conclusion
The Ph- ALL extramedullary relapses show higher genetic instability, with similar low frequency mutation profile. WES suggest that JAK2 alterations are driver mutations. This finding could be helpful for a targeted therapy. Supported by: ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: E-poster
Keyword(s): Acute lymphoblastic leukemia
{{ help_message }}
{{filter}}


