CHROMOTHRIPSIS: A NEW MECHANISM OF CANCER INITIATION AND PROGRESSION IN ADULT ACUTE MYELOID LEUKEMIA
(Abstract release date: 05/19/16)
EHA Library. Fontana M. 06/10/16; 133171; P183

Maria Chiara Fontana
Contributions
Contributions
Abstract
Abstract: P183
Type: Poster Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 17:15 - 18:45
Location: Poster area (Hall H)
Background
Chromosomal abnormalities are predictive of response in Acute Myeloid Leukemia (AML) and guide therapeutic strategies. Chromothripsis, a one-step catastrophic mechanism of genomic instability, could represent a driving force in the development and progression of hematological diseases.
Aims
To discover the mechanisms underlying the pathogenesis, chromosomal instability and heterogeneity of AML, we used single-nucleotide polymorphism (SNP) microarrays to study chromothripsis in our cohort of patients (pts).
Methods
We performed classical cytogenetic karyotyping and microarray analysis using Genome-Wide Human SNP Arrays 6.0 or Cytoscan HD Arrays (Affymetrix) in 303 AML pts at diagnosis (both de novo and secondary). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery, v.7.5). The survival analysis was performed with Kaplan-Meier method using Mantel-Cox test.
Results
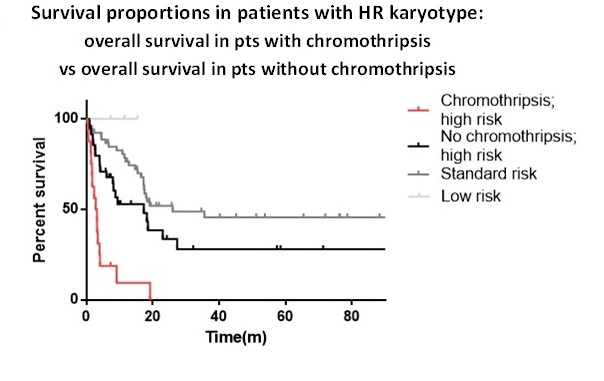
Twenty-three out of 303 pts (7.6%) showed chromothripsis events involving different chromosomes (2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 15, 16, 17 and 20), reflecting the random distribution of the one-step catastrophic event. By Nexus Copy Number we defined chromothripsis as a 2-3 variation of copy number (CN) state with at least 5 subsequent rearrangements, including CN gain and loss, interspersed with regions of diploid state and variable number of breakpoints.The pts affected by chromotripsis had a median age of 67.5 years, complex karyotype and high risk (HR) disease according to ELN definition. This group was screened for FLT3, NPM1, IDH1, IDH2 and TP53 mutational status. TP53 was evaluated in 21/23 pts, displaying that the majority (17/21) of the pts with chromothripsis harbored a TP53 mutation, while 4/21 pts were wild type for TP53 but 2 of those had a single-copy loss of the gene. TP53 mutation has a significantly higher incidence in chromothripsis pts than in others without chromothripsis (p<0.001). Chromothripsis defines a group of pts with poor prognosis compared with other in the cohort (p<0.001), with a median survival of 2.9 months and 19.1 months respectively. Remarkably, chromothripsis defines the group with the worst prognosis even if compared with pts harboring HR[JM1] karyotype features without chromothripsis (p<0.001) (Fig.1). Median survival was 2.9 months in pts with chromothripsis, 17.3 months in HR pts without chromothripsis, and 26.0 months for pts with standard risk features. Moreover, by comparing pts with (23/303) and without (280/303) chromothripsis, we identified several genes differentially altered between the 2 groups (p<0.001). The statistically most common deleted genes in chromothripsis are RAD50, MARCH3, PRDM6, SSBP2, CDC23, HDAC3, CHD1, TBCA, LMNB1, JMY, with 5q being the main altered chromosomic region. While the significant amplifications included ZDPM2, RUNX2, RUNX1T1, FLT3, ERG, TTC3 and GPC6. The most significantly affected pathways in the chromothripsis group of pts are: regulation and extension of axon, canonical Wnt signaling pathway involved in regulation of cell proliferation, positive regulation of mitotic cell cycle, chromatin organization, telomere formation and maintenance (p<0.001).
Conclusion
Chromothripsis is a recurrent event in adult AML and independently defines a group of pts with poor prognosis. It is strongly associated with TP53 mutations and losses, highlighting the importance of TP53 for maintaining genomic stability in adult AML.Acknowledgements: ELN, AIL, AIRC, PRIN, Progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.
Session topic: Acute myeloid leukemia - Biology 1
Keyword(s): Acute leukemia, Chromosomal instability
Type: Poster Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 17:15 - 18:45
Location: Poster area (Hall H)
Background
Chromosomal abnormalities are predictive of response in Acute Myeloid Leukemia (AML) and guide therapeutic strategies. Chromothripsis, a one-step catastrophic mechanism of genomic instability, could represent a driving force in the development and progression of hematological diseases.
Aims
To discover the mechanisms underlying the pathogenesis, chromosomal instability and heterogeneity of AML, we used single-nucleotide polymorphism (SNP) microarrays to study chromothripsis in our cohort of patients (pts).
Methods
We performed classical cytogenetic karyotyping and microarray analysis using Genome-Wide Human SNP Arrays 6.0 or Cytoscan HD Arrays (Affymetrix) in 303 AML pts at diagnosis (both de novo and secondary). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery, v.7.5). The survival analysis was performed with Kaplan-Meier method using Mantel-Cox test.
Results
Twenty-three out of 303 pts (7.6%) showed chromothripsis events involving different chromosomes (2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 15, 16, 17 and 20), reflecting the random distribution of the one-step catastrophic event. By Nexus Copy Number we defined chromothripsis as a 2-3 variation of copy number (CN) state with at least 5 subsequent rearrangements, including CN gain and loss, interspersed with regions of diploid state and variable number of breakpoints.The pts affected by chromotripsis had a median age of 67.5 years, complex karyotype and high risk (HR) disease according to ELN definition. This group was screened for FLT3, NPM1, IDH1, IDH2 and TP53 mutational status. TP53 was evaluated in 21/23 pts, displaying that the majority (17/21) of the pts with chromothripsis harbored a TP53 mutation, while 4/21 pts were wild type for TP53 but 2 of those had a single-copy loss of the gene. TP53 mutation has a significantly higher incidence in chromothripsis pts than in others without chromothripsis (p<0.001). Chromothripsis defines a group of pts with poor prognosis compared with other in the cohort (p<0.001), with a median survival of 2.9 months and 19.1 months respectively. Remarkably, chromothripsis defines the group with the worst prognosis even if compared with pts harboring HR[JM1] karyotype features without chromothripsis (p<0.001) (Fig.1). Median survival was 2.9 months in pts with chromothripsis, 17.3 months in HR pts without chromothripsis, and 26.0 months for pts with standard risk features. Moreover, by comparing pts with (23/303) and without (280/303) chromothripsis, we identified several genes differentially altered between the 2 groups (p<0.001). The statistically most common deleted genes in chromothripsis are RAD50, MARCH3, PRDM6, SSBP2, CDC23, HDAC3, CHD1, TBCA, LMNB1, JMY, with 5q being the main altered chromosomic region. While the significant amplifications included ZDPM2, RUNX2, RUNX1T1, FLT3, ERG, TTC3 and GPC6. The most significantly affected pathways in the chromothripsis group of pts are: regulation and extension of axon, canonical Wnt signaling pathway involved in regulation of cell proliferation, positive regulation of mitotic cell cycle, chromatin organization, telomere formation and maintenance (p<0.001).
Conclusion
Chromothripsis is a recurrent event in adult AML and independently defines a group of pts with poor prognosis. It is strongly associated with TP53 mutations and losses, highlighting the importance of TP53 for maintaining genomic stability in adult AML.Acknowledgements: ELN, AIL, AIRC, PRIN, Progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: Acute myeloid leukemia - Biology 1
Keyword(s): Acute leukemia, Chromosomal instability
Abstract: P183
Type: Poster Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 17:15 - 18:45
Location: Poster area (Hall H)
Background
Chromosomal abnormalities are predictive of response in Acute Myeloid Leukemia (AML) and guide therapeutic strategies. Chromothripsis, a one-step catastrophic mechanism of genomic instability, could represent a driving force in the development and progression of hematological diseases.
Aims
To discover the mechanisms underlying the pathogenesis, chromosomal instability and heterogeneity of AML, we used single-nucleotide polymorphism (SNP) microarrays to study chromothripsis in our cohort of patients (pts).
Methods
We performed classical cytogenetic karyotyping and microarray analysis using Genome-Wide Human SNP Arrays 6.0 or Cytoscan HD Arrays (Affymetrix) in 303 AML pts at diagnosis (both de novo and secondary). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery, v.7.5). The survival analysis was performed with Kaplan-Meier method using Mantel-Cox test.
Results
Twenty-three out of 303 pts (7.6%) showed chromothripsis events involving different chromosomes (2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 15, 16, 17 and 20), reflecting the random distribution of the one-step catastrophic event. By Nexus Copy Number we defined chromothripsis as a 2-3 variation of copy number (CN) state with at least 5 subsequent rearrangements, including CN gain and loss, interspersed with regions of diploid state and variable number of breakpoints.The pts affected by chromotripsis had a median age of 67.5 years, complex karyotype and high risk (HR) disease according to ELN definition. This group was screened for FLT3, NPM1, IDH1, IDH2 and TP53 mutational status. TP53 was evaluated in 21/23 pts, displaying that the majority (17/21) of the pts with chromothripsis harbored a TP53 mutation, while 4/21 pts were wild type for TP53 but 2 of those had a single-copy loss of the gene. TP53 mutation has a significantly higher incidence in chromothripsis pts than in others without chromothripsis (p<0.001). Chromothripsis defines a group of pts with poor prognosis compared with other in the cohort (p<0.001), with a median survival of 2.9 months and 19.1 months respectively. Remarkably, chromothripsis defines the group with the worst prognosis even if compared with pts harboring HR[JM1] karyotype features without chromothripsis (p<0.001) (Fig.1). Median survival was 2.9 months in pts with chromothripsis, 17.3 months in HR pts without chromothripsis, and 26.0 months for pts with standard risk features. Moreover, by comparing pts with (23/303) and without (280/303) chromothripsis, we identified several genes differentially altered between the 2 groups (p<0.001). The statistically most common deleted genes in chromothripsis are RAD50, MARCH3, PRDM6, SSBP2, CDC23, HDAC3, CHD1, TBCA, LMNB1, JMY, with 5q being the main altered chromosomic region. While the significant amplifications included ZDPM2, RUNX2, RUNX1T1, FLT3, ERG, TTC3 and GPC6. The most significantly affected pathways in the chromothripsis group of pts are: regulation and extension of axon, canonical Wnt signaling pathway involved in regulation of cell proliferation, positive regulation of mitotic cell cycle, chromatin organization, telomere formation and maintenance (p<0.001).
Conclusion
Chromothripsis is a recurrent event in adult AML and independently defines a group of pts with poor prognosis. It is strongly associated with TP53 mutations and losses, highlighting the importance of TP53 for maintaining genomic stability in adult AML.Acknowledgements: ELN, AIL, AIRC, PRIN, Progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: Acute myeloid leukemia - Biology 1
Keyword(s): Acute leukemia, Chromosomal instability
Type: Poster Presentation
Presentation during EHA21: On Friday, June 10, 2016 from 17:15 - 18:45
Location: Poster area (Hall H)
Background
Chromosomal abnormalities are predictive of response in Acute Myeloid Leukemia (AML) and guide therapeutic strategies. Chromothripsis, a one-step catastrophic mechanism of genomic instability, could represent a driving force in the development and progression of hematological diseases.
Aims
To discover the mechanisms underlying the pathogenesis, chromosomal instability and heterogeneity of AML, we used single-nucleotide polymorphism (SNP) microarrays to study chromothripsis in our cohort of patients (pts).
Methods
We performed classical cytogenetic karyotyping and microarray analysis using Genome-Wide Human SNP Arrays 6.0 or Cytoscan HD Arrays (Affymetrix) in 303 AML pts at diagnosis (both de novo and secondary). SNP Array data were analyzed by Nexus Copy Number™ v7.5 (BioDiscovery, v.7.5). The survival analysis was performed with Kaplan-Meier method using Mantel-Cox test.
Results
Twenty-three out of 303 pts (7.6%) showed chromothripsis events involving different chromosomes (2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 15, 16, 17 and 20), reflecting the random distribution of the one-step catastrophic event. By Nexus Copy Number we defined chromothripsis as a 2-3 variation of copy number (CN) state with at least 5 subsequent rearrangements, including CN gain and loss, interspersed with regions of diploid state and variable number of breakpoints.The pts affected by chromotripsis had a median age of 67.5 years, complex karyotype and high risk (HR) disease according to ELN definition. This group was screened for FLT3, NPM1, IDH1, IDH2 and TP53 mutational status. TP53 was evaluated in 21/23 pts, displaying that the majority (17/21) of the pts with chromothripsis harbored a TP53 mutation, while 4/21 pts were wild type for TP53 but 2 of those had a single-copy loss of the gene. TP53 mutation has a significantly higher incidence in chromothripsis pts than in others without chromothripsis (p<0.001). Chromothripsis defines a group of pts with poor prognosis compared with other in the cohort (p<0.001), with a median survival of 2.9 months and 19.1 months respectively. Remarkably, chromothripsis defines the group with the worst prognosis even if compared with pts harboring HR[JM1] karyotype features without chromothripsis (p<0.001) (Fig.1). Median survival was 2.9 months in pts with chromothripsis, 17.3 months in HR pts without chromothripsis, and 26.0 months for pts with standard risk features. Moreover, by comparing pts with (23/303) and without (280/303) chromothripsis, we identified several genes differentially altered between the 2 groups (p<0.001). The statistically most common deleted genes in chromothripsis are RAD50, MARCH3, PRDM6, SSBP2, CDC23, HDAC3, CHD1, TBCA, LMNB1, JMY, with 5q being the main altered chromosomic region. While the significant amplifications included ZDPM2, RUNX2, RUNX1T1, FLT3, ERG, TTC3 and GPC6. The most significantly affected pathways in the chromothripsis group of pts are: regulation and extension of axon, canonical Wnt signaling pathway involved in regulation of cell proliferation, positive regulation of mitotic cell cycle, chromatin organization, telomere formation and maintenance (p<0.001).
Conclusion
Chromothripsis is a recurrent event in adult AML and independently defines a group of pts with poor prognosis. It is strongly associated with TP53 mutations and losses, highlighting the importance of TP53 for maintaining genomic stability in adult AML.Acknowledgements: ELN, AIL, AIRC, PRIN, Progetto Regione-Università 2010-12 (L. Bolondi), FP7 NGS-PTL project.

Session topic: Acute myeloid leukemia - Biology 1
Keyword(s): Acute leukemia, Chromosomal instability
{{ help_message }}
{{filter}}


